6U51
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) Complexed with Sudan ebolavirus Nucleoprotein C-terminal Domain 610-738 | | Descriptor: | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB), Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
4WJ5
 
 | | Structure of HLA-A2 in complex with an altered peptide ligands based on Mart-1 variant epitope | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Celie, P.H.N, Rodenko, B, Ovaa, H. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Altered Peptide Ligands Revisited: Vaccine Design through Chemically Modified HLA-A2-Restricted T Cell Epitopes.
J Immunol., 193, 2014
|
|
4WGF
 
 | | YcaC from Pseudomonas aeruginosa with hexane-2,5-diol and covalent acrylamide | | Descriptor: | (2R,5R)-hexane-2,5-diol, CHLORIDE ION, PROPIONAMIDE, ... | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34020686 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
3NRZ
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Hypoxanthine | | Descriptor: | DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cao, H, Pauff, J.M, Hille, R. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid.
J.Biol.Chem., 285, 2010
|
|
3NVV
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Arsenite | | Descriptor: | ARSENITE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structure of Arsenite-Inhibited Xanthine Oxidase: Mu-Sulfido,Mu-Oxo Double Bridge between Molybdenum and Arsenic in the Active Site.
J.Am.Chem.Soc., 133, 2011
|
|
8JCH
 
 | |
7QI2
 
 | |
8IPB
 
 | | Wheat 80S ribosome pausing on AUG-Stop with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
7OHU
 
 | |
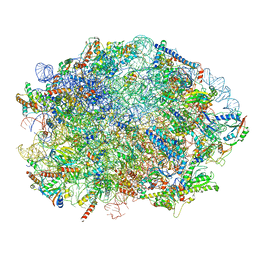
7OF1
 
 | |
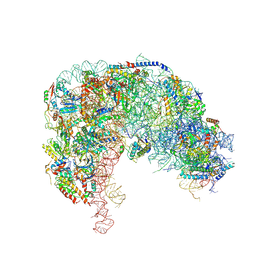
7OHY
 
 | |
3P5J
 
 | |
8K5P
 
 | |
2W8L
 
 | | Y-family DNA polymerase Dpo4 bypassing N2-naphthyl-guanine adduct in anti orientation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*CP*N2GP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Zhang, H, Egli, M, Guengerich, F.P. | | Deposit date: | 2009-01-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Versatility of Y-Family Sulfolobus Solfataricus DNA Polymerase Dpo4 in Translesion Synthesis Past Bulky N2-Alkylguanine Adducts.
J.Biol.Chem., 284, 2009
|
|
2W8K
 
 | | Y-family DNA polymerase Dpo4 bypassing N2-naphthyl-guanine adduct in syn orientation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*CP*N2GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Zhang, H, Egli, M, Guengerich, F.P. | | Deposit date: | 2009-01-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Versatility of Y-Family Sulfolobus Solfataricus DNA Polymerase Dpo4 in Translesion Synthesis Past Bulky N2-Alkylguanine Adducts.
J.Biol.Chem., 284, 2009
|
|
6VY9
 
 | | Crystal structure of NotF prenyltransferase | | Descriptor: | Deoxybrevianamide E synthase notF | | Authors: | Dan, Q, Smith, J.L. | | Deposit date: | 2020-02-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Data Science-Driven Analysis of Substrate-Permissive Diketopiperazine Reverse Prenyltransferase NotF: Applications in Protein Engineering and Cascade Biocatalytic Synthesis of (-)-Eurotiumin A.
J.Am.Chem.Soc., 144, 2022
|
|
3MAV
 
 | | Crystal structure of Plasmodium vivax putative farnesyl pyrophosphate synthase (Pv092040) | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION | | Authors: | Dong, A, Dunford, J, Lew, J, Wernimont, A.K, Ren, H, Zhao, Y, Koeieradzki, I, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
4OKQ
 
 | |
3D9S
 
 | | Human Aquaporin 5 (AQP5) - High Resolution X-ray Structure | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Horsefield, R, Norden, K, Fellert, M, Backmark, A, Tornroth-Horsefield, S, Terwisscha Van Scheltinga, A.C, Kvassman, J, Kjellbom, P, Johanson, U, Neutze, R. | | Deposit date: | 2008-05-27 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution x-ray structure of human aquaporin 5
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3N8W
 
 | |
3N8Y
 
 | | Structure of Aspirin Acetylated Cyclooxygenase-1 in Complex with Diclofenac | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3N8Z
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3NS1
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with 6-Mercaptopurine | | Descriptor: | 9H-purine-6-thiol, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Pauff, J.M, Hille, R. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid.
J.Biol.Chem., 285, 2010
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
