6POM
 
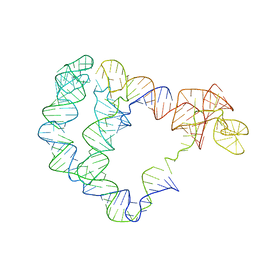 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6SPF
 
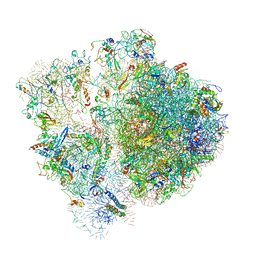 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPD
 
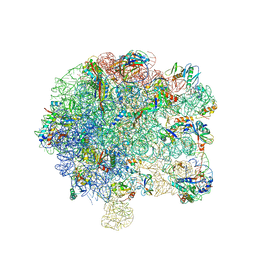 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPE
 
 | | Pseudomonas aeruginosa 30s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4TUA
 
 | |
4TUC
 
 | |
4TUE
 
 | |
4TUD
 
 | |
6SPB
 
 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate with a mutation in uL6 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4W7S
 
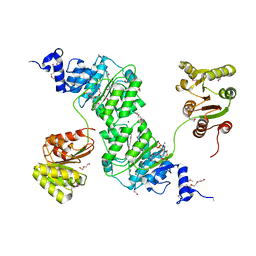 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
1LU3
 
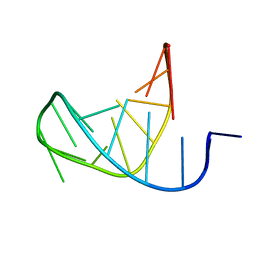 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | PHENYLALANINE TRANSFER RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
5ZET
 
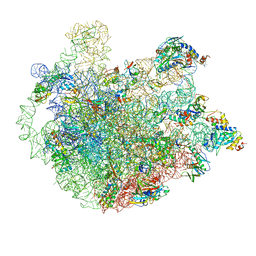 | | M. smegmatis P/P state 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
8BWS
 
 | | Structure of yeast RNA Polymerase III elongation complex at 3.3 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
6HCT
 
 | |
5ZAK
 
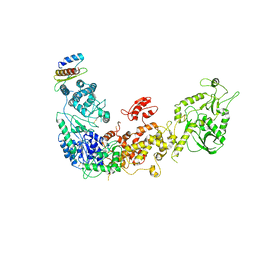 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
7R97
 
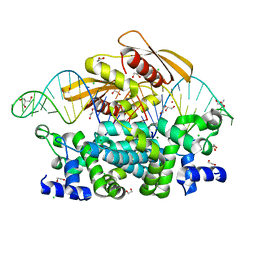 | | Crystal structure of postcleavge complex of Escherichia coli RNase III | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Dharavath, S, Shaw, G.X, Ji, X. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg 2+ -ion catalysis.
Rna Biol., 19, 2022
|
|
8PV4
 
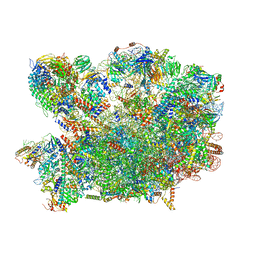 | | Chaetomium thermophilum pre-60S State 2 - pre-5S rotation with Rix1 complex - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
7Z2Z
 
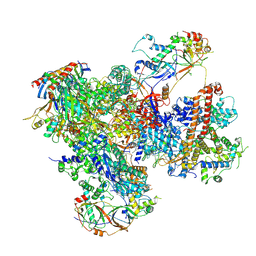 | | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
8PV6
 
 | | Chaetomium thermophilum pre-60S State 3 - post-5S rotation with Rix1 complex with Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
7NQ4
 
 | |
7S3C
 
 | |
7S3A
 
 | |
7OQE
 
 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
