6GV1
 
 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
4JQ8
 
 | | Crystal structure of EGFR kinase domain in complex with compound 4b | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]-N~3~,N~3~-dimethyl-beta-alaninamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JR3
 
 | | Crystal structure of EGFR kinase domain in complex with compound 3g | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]acetamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4N0H
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4N0I
 
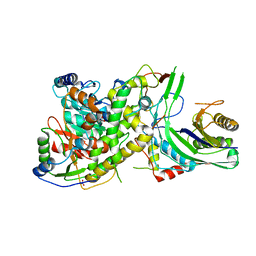 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
7V86
 
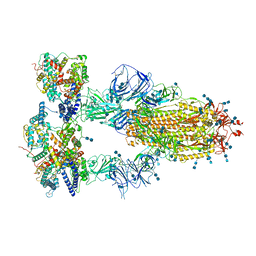 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V81
 
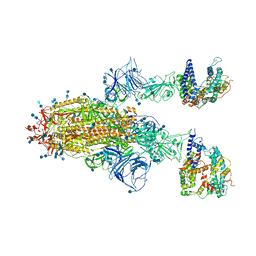 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form
To Be Published
|
|
7V83
 
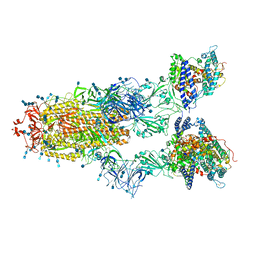 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
To Be Published
|
|
7V84
 
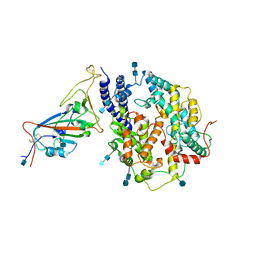 | | Local refinement of SARS-CoV-2 S-Gamma variant (P.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Gamma variant (P.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7V85
 
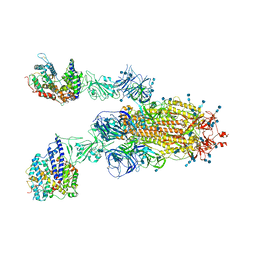 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form
To Be Published
|
|
6D90
 
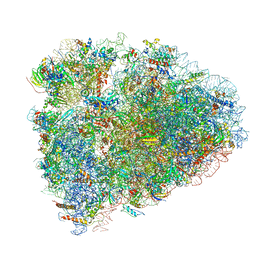 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-site tRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-27 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
6T36
 
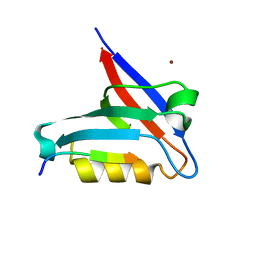 | | Crystal structure of the PTPN3 PDZ domain bound to the HBV core protein C-terminal peptide | | Descriptor: | BROMIDE ION, Capsid protein, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | Deposit date: | 2019-10-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein.
Sci Rep, 11, 2021
|
|
4NP6
 
 | | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor | | Descriptor: | Adenylate kinase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor
To be Published
|
|
4NH1
 
 | | Crystal structure of a heterotetrameric CK2 holoenzyme complex carrying the Andante-mutation in CK2beta and consistent with proposed models of autoinhibition and trans-autophosphorylation | | Descriptor: | Casein kinase II subunit alpha, Casein kinase II subunit beta, GLYCEROL, ... | | Authors: | Schnitzler, A, Issinger, O.-G, Niefind, K. | | Deposit date: | 2013-11-04 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Protein Kinase CK2(Andante) Holoenzyme Structure Supports Proposed Models of Autoregulation and Trans-Autophosphorylation
J.Mol.Biol., 426, 2014
|
|
6T83
 
 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7T
 
 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
10MH
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
1AK2
 
 | | ADENYLATE KINASE ISOENZYME-2 | | Descriptor: | ADENYLATE KINASE ISOENZYME-2, SULFATE ION | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of bovine mitochondrial adenylate kinase: comparison with isoenzymes in other compartments.
Protein Sci., 5, 1996
|
|
1ANK
 
 | | THE CLOSED CONFORMATION OF A HIGHLY FLEXIBLE PROTEIN: THE STRUCTURE OF E. COLI ADENYLATE KINASE WITH BOUND AMP AND AMPPNP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Berry, M.B, Meador, B, Bilderback, T, Liang, P, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP.
Proteins, 19, 1994
|
|
1AKY
 
 | |
1AKE
 
 | |
2EKC
 
 | |
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
7R81
 
 | | Structure of the translating Neurospora crassa ribosome arrested by cycloheximide | | Descriptor: | 18S rRNA, 26S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Shen, L, Su, Z, Yang, K, Wu, C, Becker, T, Bell-Pedersen, D, Zhang, J, Sachs, M.S. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the translating Neurospora ribosome arrested by cycloheximide
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
