8QE4
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QOR
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QDU
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
4GOL
 
 | |
1KX3
 
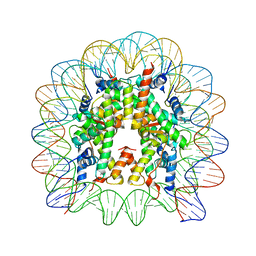 | | X-Ray Structure of the Nucleosome Core Particle, NCP146, at 2.0 A Resolution | | Descriptor: | DNA (5'(ATCAATATCCACCTGCAGATTCTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGAATTCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGAATCTGCAGGTGGATATTGAT)3'), MANGANESE (II) ION, histone H2A.1, ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
4GBE
 
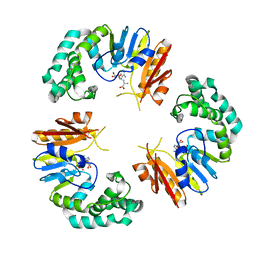 | |
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4WU9
 
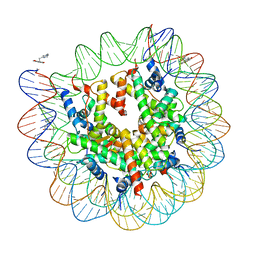 | | Structure of cisPtNAP-NCP145 | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Davey, G.E, Chin, C.F, Droge, P, Ang, W.H, Davey, C.A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stereochemical control of nucleosome targeting by platinum-intercalator antitumor agents.
Nucleic Acids Res., 43, 2015
|
|
8DJQ
 
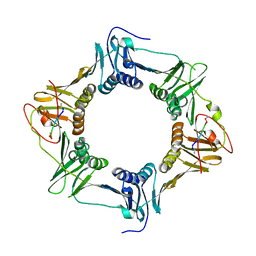 | | Sliding-clamp-DnaE1 peptide | | Descriptor: | ACETYL GROUP, AMINO GROUP, Beta sliding clamp, ... | | Authors: | Kapur, M.K, Gray, O.J, Honzatko, R.H, Nelson, S.N. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interaction of the sliding clamp with mycobacterial polymerases
To Be Published
|
|
4WU8
 
 | | Structure of trPtNAP-NCP145 | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Stereochemical control of nucleosome targeting by platinum-intercalator antitumor agents.
Nucleic Acids Res., 43, 2015
|
|
8BJM
 
 | |
169D
 
 | | THE SOLUTION STRUCTURE OF THE R(GCG)D(TATACCC):D(GGGTATACGC) OKAZAKI FRAGMENT CONTAINS TWO DISTINCT DUPLEX MORPHOLOGIES CONNECTED BY A JUNCTION | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*TP*AP*CP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*G)-D(P*TP*AP*TP*AP*CP*CP*C)-3') | | Authors: | Salazar, M, Fedoroff, O.Y, Zhu, L, Reid, B.R. | | Deposit date: | 1994-04-11 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction.
J.Mol.Biol., 241, 1994
|
|
2M9H
 
 | |
3I7L
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of DDB2 | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8HR5
 
 | | Cryo-EM structure of the CnCas12f1-sgRNA-DNA complex | | Descriptor: | DNA (28-MER), DNA (5'-D(*TP*AP*AP*CP*CP*TP*AP*AP*TP*AP*GP*AP*TP*GP*TP*GP*AP*A)-3'), Transposase, ... | | Authors: | Li, F, Ji, Q. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure and engineering of miniature Clostridium novyi CRISPR-Cas12f1 with rare C-rich PAM specificity
To Be Published
|
|
2A6H
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic sterptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Temiakov, D, Zenkin, N, Vassylyeva, M.N, Perederina, A, Tahirov, T.H, Savkina, M, Zorov, S, Nikiforov, V, Igarashi, N, Matsugaki, N, Wakatsuki, S, Severinov, K, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of transcription inhibition by antibiotic streptolydigin.
Mol.Cell, 19, 2005
|
|
2E9X
 
 | | The crystal structure of human GINS core complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Kamada, K, Hanaoka, F. | | Deposit date: | 2007-01-27 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human GINS complex and its assembly and functional interface in replication initiation
Nat.Struct.Mol.Biol., 14, 2007
|
|
9BDC
 
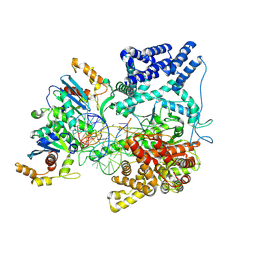 | | Cryo-EM Structure of the TEFM bound Human Mitochondrial Transcription Elongation Complex in a Closed Fingers Domain Conformation | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Non-Template Strand DNA (NT27mt_+1T), ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
2IX2
 
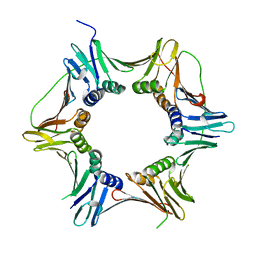 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5IP3
 
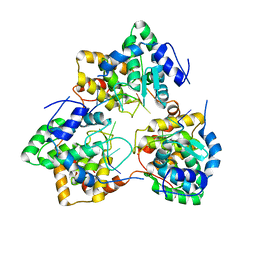 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssDNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
7AY1
 
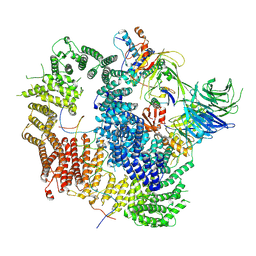 | | Cryo-EM structure of USP1-UAF1 bound to mono-ubiquitinated FANCD2, and FANCI | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Rennie, M.L, Arkinson, C, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OTQ
 
 | | Cryo-EM structure of ALC1/CHD1L bound to a PARylated nucleosome | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome
Elife, 10, 2021
|
|
8U3K
 
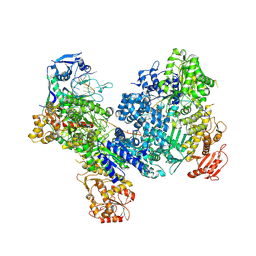 | | DdmDE handover complex | | Descriptor: | DNA (29-MER), DNA (5'-D(P*GP*GP*AP*AP*AP*TP*GP*TP*TP*GP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
6TEY
 
 | |
150D
 
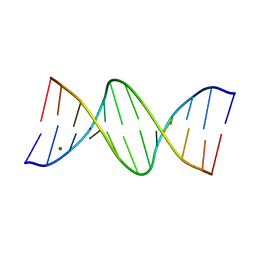 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
