4I5K
 
 | |
6HDT
 
 | |
5A7B
 
 | |
3LZW
 
 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from bacillus subtilis (form I) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
6HDS
 
 | | Crystal Structure of apo short afifavidin | | Descriptor: | short afifavidin | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2018-08-19 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of afifavidin reveals common features of molecular assemblage in the bacterial dimeric avidins.
FEBS J., 285, 2018
|
|
7K1Q
 
 | |
3GC9
 
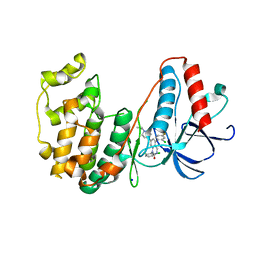 | | The structure of p38beta C119S, C162S in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, SODIUM ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4C2K
 
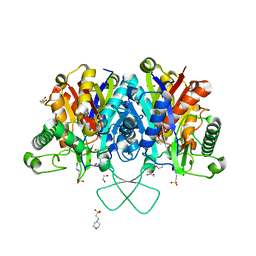 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5N5H
 
 | | Crystal structure of metallo-beta-lactamase VIM-1 in complex with ML302F inhibitor | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
3LZX
 
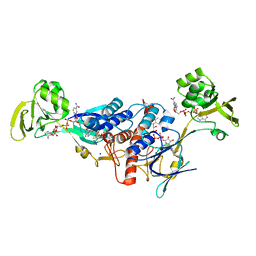 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from Bacillus subtilis (FORM II) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
5TMC
 
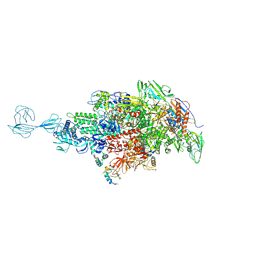 | | Re-refinement of Thermus thermopiles DNA-directed RNA polymerase structure | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
3N2C
 
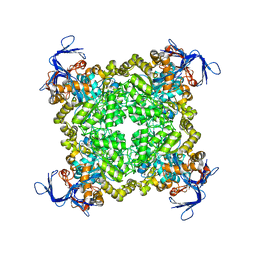 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
1JT9
 
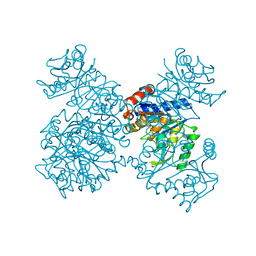 | | Structure of the mutant F174A T form of the Glucosamine-6-Phosphate deaminase from E.coli | | Descriptor: | Glucosamine-6-Phosphate deaminase | | Authors: | Bustos-Jaimes, I, Sosa-Peinado, A, Rudino-Pinera, E, Horjales, E, Calcagno, M.L. | | Deposit date: | 2001-08-20 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the conformational flexibility of the active-site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase.
J.Mol.Biol., 319, 2002
|
|
3GM7
 
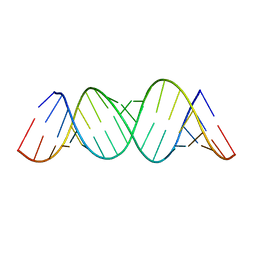 | |
1GK7
 
 | | HUMAN VIMENTIN COIL 1A FRAGMENT (1A) | | Descriptor: | SULFATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
4C2J
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6K35
 
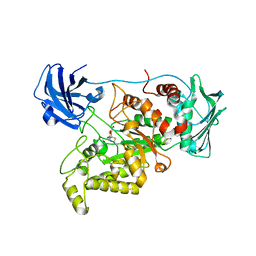 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
3ZD4
 
 | |
5BPX
 
 | | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP. | | Descriptor: | 2,4'-dihydroxyacetophenone dioxygenase, ACETATE ION, FE (III) ION, ... | | Authors: | Guo, J, Erskine, P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Extension of resolution and oligomerization-state studies of 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3MTW
 
 | | Crystal structure of L-Lysine, L-Arginine carboxypeptidase Cc2672 from Caulobacter Crescentus CB15 complexed with N-methyl phosphonate derivative of L-Arginine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L-Arginine carboxypeptidase Cc2672, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-05-01 | | Release date: | 2010-07-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Identification and Structure Determination of Two Novel Prolidases from cog1228 in the Amidohydrolase Superfamily
Biochemistry, 49, 2010
|
|
1Q02
 
 | | NMR structure of the UBA domain of p62 (SQSTM1) | | Descriptor: | sequestosome 1 | | Authors: | Ciani, B, Layfield, R, Cavey, J.R, Sheppard, P.W, Searle, M.S. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-associated Domain of p62 (SQSTM1) and Implications for Mutations That Cause Paget's
Disease of Bone
J.Biol.Chem., 278, 2003
|
|
4MXR
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase with Mn2+2 | | Descriptor: | Formiminoglutamase, GLYCEROL, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.-J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
4MYN
 
 | | Crystal structure of Trypanosoma cruzi formiminoglutamase N114H variant with Mn2+2 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
