1MSS
 
 | |
5F2G
 
 | |
5F4W
 
 | |
7C9N
 
 | | Crystal structure of SETDB1 tudor domain in complexed with Compound 1. | | Descriptor: | 3,5-dimethyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y, Xiong, L, Mao, X, Yang, S. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1U8Q
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELEKWAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1U93
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog EQDKW-[Dap]-S (cyclic) | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE ANALOG | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
5V8Q
 
 | | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs): Part III | | Descriptor: | 4-[(2S,3S)-2-ethyl-3-hydroxy-5-oxopyrrolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, GLYCEROL | | Authors: | Wilson, K.P. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs) Part III: Discovery of 4-(5-oxopyrrolidine-1-yl)benzonitrile derivative 2f as a clinical candidate.
Bioorg. Med. Chem., 25, 2017
|
|
5FEQ
 
 | | EGFR KINASE DOMAIN IN COMPLEX WITH A COVALENT AMINOBENZIMIDAZOLE | | Descriptor: | Epidermal growth factor receptor, ~{N}-[1-[(3~{R})-1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl]-7-methyl-benzimidazol-2-yl]-2-methyl-pyridine-4-carboxamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
2RLH
 
 | |
5FB8
 
 | | Structure of Interleukin-16 bound to the 14.1 antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-IL-16 antibody 14.1 Fab domain Heavy Chain, ... | | Authors: | Hall, G, Cowan, R, Bayliss, R, Carr, M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a Potential Therapeutic Antibody Bound to Interleukin-16 (IL-16): MECHANISTIC INSIGHTS AND NEW THERAPEUTIC OPPORTUNITIES.
J.Biol.Chem., 291, 2016
|
|
1U8I
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWAN | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
4OVL
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E)-(2-oxidanylidene-1H-indol-3-ylidene)methyl]benzoic acid, ACETIC ACID, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV integrase for compounds that bind--a SAMPL challenge.
J. Comput. Aided Mol. Des., 28, 2014
|
|
4RIO
 
 | |
1MHP
 
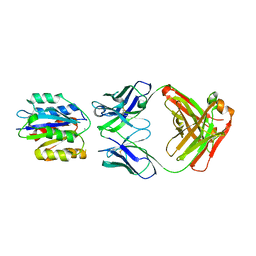 | | Crystal structure of a chimeric alpha1 integrin I-domain in complex with the Fab fragment of a humanized neutralizing antibody | | Descriptor: | FAB FRAGMENT, light chain, Fab fragment, ... | | Authors: | Karpusas, M, Taylor, F, Ferrant, J, Weinreb, P, Garber, E. | | Deposit date: | 2002-08-20 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the alpha 1 beta 1 Integrin I Domain in Complex with
an Antibody Fab Fragment
J.Mol.Biol., 327, 2003
|
|
4CDM
 
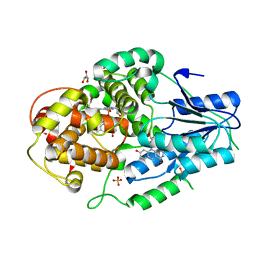 | | Crystal structure of M. mazei photolyase soaked with synthetic 8-HDF | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
3TUX
 
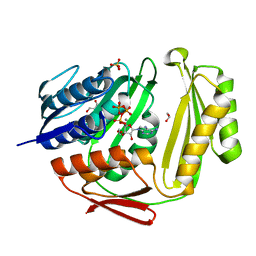 | | Crystal structure of RtcA.ATP.Mn ternary complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2011-09-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of RNA 3'-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KR3
 
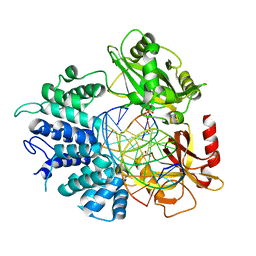 | |
2R1M
 
 | | OpdA from Agrobacterium radiobacter with bound product diethyl phosphate from crystal soaking with diethyl 4-methoxyphenyl phosphate (450h)- 2.5 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
4MTI
 
 | | Crystal structure of cIAP1 BIR3 bound to T3258042 | | Descriptor: | (3S,8aR)-2-[(2S)-2-cyclohexyl-2-{[(2S)-2-(methylamino)butanoyl]amino}acetyl]-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Snell, G.P, Dougan, D.R. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, synthesis, and biological activities of novel hexahydropyrazino[1,2-a]indole derivatives as potent inhibitors of apoptosis (IAP) proteins antagonists with improved membrane permeability across MDR1 expressing cells.
Bioorg.Med.Chem., 21, 2013
|
|
4TM7
 
 | | Crystal structure of 6-phosphogluconolactonase from Mycobacterium smegmatis N131D mutant soaked with CuSO4 | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphogluconolactonase, CHLORIDE ION, ... | | Authors: | Fujieda, N, Stuttfeld, E, Maier, T. | | Deposit date: | 2014-05-31 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Enzyme repurposing of a hydrolase as an emergent peroxidase upon metal binding.
Chem Sci, 6, 2015
|
|
7KR4
 
 | |
7DU1
 
 | |
7DUE
 
 | |
4OGG
 
 | |
