6TVI
 
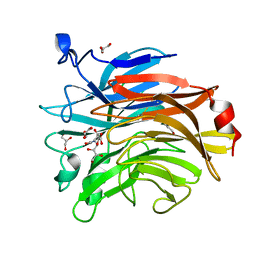 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
5ZGE
 
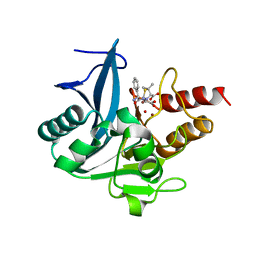 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
3GHR
 
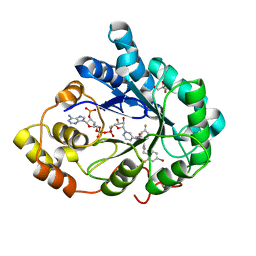 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. First stage of radiation damage | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
5M2K
 
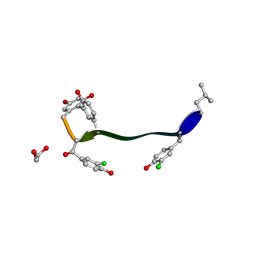 | | Crystal structure of vancomycin-Zn(II) complex | | Descriptor: | 1,2-ETHANEDIOL, ZINC ION, vancomycin, ... | | Authors: | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
4X6H
 
 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | Descriptor: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | Authors: | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
5E1K
 
 | | Selenomethionine Ca2+-Calmodulin from Paramecium tetraurelia SAD data | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2AT8
 
 | |
4B4E
 
 | | 1.00 A Structure of Lysozyme Crystallized with (R)-2-methyl-2,4- pentanediol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, LYSOZYME C | | Authors: | Jakoncic, J, Berger, J, Stauber, M, Axelbaum, A, Asherie, N. | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallization of Lysozyme with (R)-, (S)- and (Rs)-2-Methyl-2,4-Pentanediol
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1K4P
 
 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with zinc ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, SULFATE ION, ZINC ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
3HYD
 
 | |
4TXR
 
 | |
1NQJ
 
 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.0 ANGSTROM RESOLUTION IN ABSENCE OF CALCIUM | | Descriptor: | CHLORIDE ION, LITHIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A bacterial collagen-binding domain with novel calcium-binding motif
controls domain orientation
Embo J., 22, 2003
|
|
5WHP
 
 | | Crystal structure of the segment, NFGTFS, from the A315T familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6QLN
 
 | | Galectin-3C in complex with fluoroaryl triazole monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
8TSX
 
 | |
3E7R
 
 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3I34
 
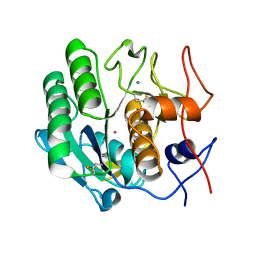 | | Proteinase K by LB Nanotemplate Method after high X-Ray dose on ID14-2 Beamline at ESRF | | Descriptor: | CALCIUM ION, MERCURY (II) ION, Proteinase K | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-06-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Radiation damage study of Proteinase K at ID14-2 beamline at ESRF
To be Published
|
|
7FW9
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-ethoxycarbonyl-4,5,6,7-tetrahydro-1-benzothiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-{[3-(ethoxycarbonyl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
5RS9
 
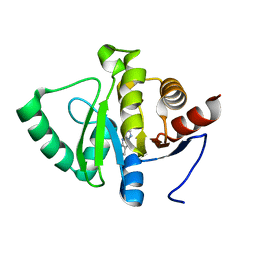 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000007636250 | | Descriptor: | 6,7-dihydro-5H-cyclopenta[d][1,2,4]triazolo[1,5-a]pyrimidin-8-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RT7
 
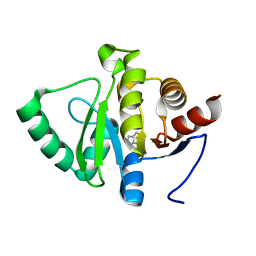 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000015442276 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTO
 
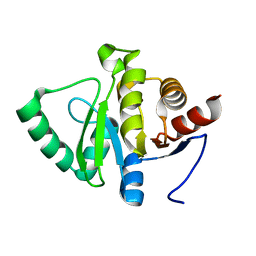 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388302 | | Descriptor: | 4-PIPERIDINO-PIPERIDINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU6
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001442764 | | Descriptor: | Non-structural protein 3, naphthalene-2-carboximidamide | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001683100 | | Descriptor: | 4-chloro-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTJ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000332752 | | Descriptor: | Non-structural protein 3, P-HYDROXYBENZOIC ACID | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTY
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000157088 | | Descriptor: | 4-HYDROXYBENZAMIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
