5RU8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000154817 | | Descriptor: | ISOQUINOLIN-1-AMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2QX3
 
 | |
5RT9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388280 | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3PLQ
 
 | | Crystal structure of PKA type I regulatory subunit bound with Rp-8-Br-cAMPS | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-(6-amino-8-bromo-9H-purin-9-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-sulfide, ZINC ION, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Swaminathan, K. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: Conformational selection highlights a new concept in allosteric inhibitor design
To be Published
|
|
5RUP
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000004976927 | | Descriptor: | Non-structural protein 3, [3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetic acid | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTP
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001679336 | | Descriptor: | 2-oxidanylidene-2-phenylazanyl-ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV6
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000158540 | | Descriptor: | 1,3-benzodioxole-5-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2H0B
 
 | | Crystal Structure of the second LNS/LG domain from Neurexin 1 alpha | | Descriptor: | CALCIUM ION, GLYCEROL, Neurexin-1-alpha | | Authors: | Sheckler, L.R, Henry, L, Sugita, S, Sudhof, T.C, Rudenko, G. | | Deposit date: | 2006-05-14 | | Release date: | 2006-06-20 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Second LNS/LG Domain from Neurexin 1{alpha}: Ca2+ binding and the effects of alternative splicing
J.Biol.Chem., 281, 2006
|
|
5RU7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000003591110 | | Descriptor: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVH
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000265642 | | Descriptor: | Non-structural protein 3, quinoline-3-carboxylic acid | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2QZW
 
 | |
2F84
 
 | |
5RUM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000008861082 | | Descriptor: | 3-(3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV3
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000057162 | | Descriptor: | (5-methoxy-1H-indol-3-yl)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2FKM
 
 | | PMM/PGM S108D mutant with alpha-d-glucose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Regni, C.A, Beamer, L.J. | | Deposit date: | 2006-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme.
J.Biol.Chem., 281, 2006
|
|
5RVM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000157088 | | Descriptor: | 4-HYDROXYBENZAMIDE, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2H0D
 
 | |
2R0R
 
 | |
2R0I
 
 | |
2H34
 
 | | Apoenzyme crystal structure of the tuberculosis serine/threonine kinase, PknE | | Descriptor: | BROMIDE ION, SODIUM ION, Serine/threonine-protein kinase pknE | | Authors: | Gay, L.M, Ng, H.L, Alber, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conserved Dimer and Global Conformational Changes in the Structure of apo-PknE Ser/Thr Protein Kinase from Mycobacterium tuberculosis.
J.Mol.Biol., 360, 2006
|
|
2H17
 
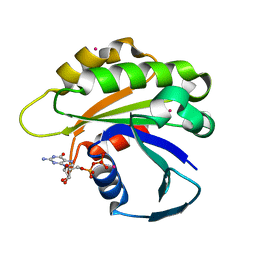 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2R1A
 
 | |
2R19
 
 | |
2H3G
 
 | |
2H1E
 
 | |
