6MYG
 
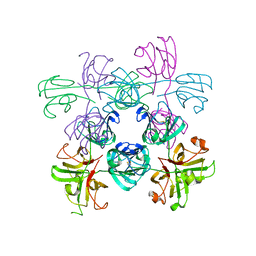 | | Mouse Gamma S Crystallin L16 Octamer | | Descriptor: | Gamma-crystallin S | | Authors: | Sagar, V, Wistow, G.J. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.919 Å) | | Cite: | Disulfides, domain swaps and disorder in a gamma-crystallin:
A model for mechanisms of aggregation in globular proteins.
To Be Published
|
|
3U0P
 
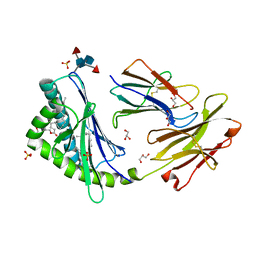 | | Crystal structure of human CD1d-lysophosphatidylcholine | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Antigen-presenting glycoprotein CD1d, Beta-2-microglobulin, ... | | Authors: | Lopez-Sagaseta, J, Sibener, L.V, Adams, E.J. | | Deposit date: | 2011-09-28 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lysophospholipid presentation by CD1d and recognition by a human Natural Killer T-cell receptor.
Embo J., 31, 2012
|
|
5GJN
 
 | |
5O14
 
 | |
1JTM
 
 | |
8Q1O
 
 | | S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 32-209), important for Self-assembly | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Sagmeister, T, Grininger, C, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5A1A
 
 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
5ANA
 
 | |
1JTN
 
 | |
1XD3
 
 | | Crystal structure of UCHL3-UbVME complex | | Descriptor: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | Authors: | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
3JR6
 
 | |
1BJG
 
 | | D221(169)N MUTANT DOES NOT PROMOTE OPENING OF THE COFACTOR IMIDAZOLIDINE RING | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Michelitsch, M.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D221 in thymidylate synthase controls conformation change, and thereby opening of the imidazolidine.
Biochemistry, 37, 1998
|
|
2B7X
 
 | |
1X41
 
 | | Solution structure of the Myb-like DNA binding domain of human Transcriptional adaptor 2-like, isoform B | | Descriptor: | Transcriptional adaptor 2-like, isoform b | | Authors: | Sasagawa, A, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Myb-like DNA binding domain of human Transcriptional adaptor 2-like, isoform B
To be Published
|
|
1X40
 
 | | Solution structure of the SAM domain of human ARAP2 | | Descriptor: | ARAP2 | | Authors: | Sasagawa, A, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM domain of human ARAP2
To be Published
|
|
3D0Z
 
 | |
3CRU
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
3CRT
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
2WWS
 
 | |
2XPJ
 
 | |
2WFP
 
 | |
1LLH
 
 | | ARE CARBOXY TERMINII OF HELICES CODED BY THE LOCAL SEQUENCE OR BY TERTIARY STRUCTURE CONTACTS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Sagermann, M, Martensson, L.-G, Baase, W.A, Matthews, B.W. | | Deposit date: | 2002-04-28 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A test of proposed rules for helix capping: Implications for protein design
Protein Sci., 11, 2002
|
|
1JQU
 
 | |
1KIA
 
 | |
2YNJ
 
 | | GroEL at sub-nanometer resolution by Constrained Single Particle Tomography | | Descriptor: | 60 KDA CHAPERONIN, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bartesaghi, A, Lecumberry, F, Sapiro, G, Subramaniam, S. | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Structure, 20, 2012
|
|
