2UYI
 
 | | Crystal structure of KSP in complex with ADP and thiophene containing inhibitor 33 | | Descriptor: | (5R)-N,N-DIETHYL-5-METHYL-2-[(THIOPHEN-2-YLCARBONYL)AMINO]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-3-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Lee, T.T. | | Deposit date: | 2007-04-07 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Sar of Thiophene Containing Kinesin Spindle Protein (Ksp) Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4W7E
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H With Copper Mediated Crystal Contacts, P 41 21 2 Space Group | | Descriptor: | COPPER (II) ION, IMIDAZOLE, fluorescent protein D21H/K26H | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7G
 
 | |
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2UZW
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.] PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2V03
 
 | | High resolution structure and catalysis of an O-acetylserine sulfhydrylase | | Descriptor: | CITRIC ACID, Cysteine synthase B, GLYCEROL, ... | | Authors: | Zocher, G, Wiesand, U, Schulz, G.E. | | Deposit date: | 2007-05-08 | | Release date: | 2007-10-09 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High resolution structure and catalysis of O-acetylserine sulfhydrylase isozyme B from Escherichia coli.
FEBS J., 274, 2007
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
4WHB
 
 | | Crystal structure of phenylurea hydrolase B | | Descriptor: | Phenylurea hydrolase B, ZINC ION | | Authors: | Sugrue, E, Carr, P.D, Khurana, J.L, Jackson, C.J. | | Deposit date: | 2014-09-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4WIB
 
 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
2UXD
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4WIH
 
 | | Crystal structure of cAMP-dependent Protein Kinase A from Cricetulus griseus | | Descriptor: | cAMP Dependent Protein Kinase Inhibitor PKI-tide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Kudlinzki, D, Linhard, V.L, Saxena, K, Dreyer, M, Schwalbe, H. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | High-resolution crystal structure of cAMP-dependent protein kinase from Cricetulus griseus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2V0W
 
 | |
2VAF
 
 | | Crystal structure of Human Cardiac Calsequestrin | | Descriptor: | CALSEQUESTRIN-2 | | Authors: | Kim, E, Youn, B, Kemper, L, Campbell, C, Milting, H, Varsanyi, M, Kang, C. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Characterization of Human Cardiac Calsequestrin and its Deleterious Mutants.
J.Mol.Biol., 373, 2007
|
|
8H53
 
 | | Human asparaginyl-tRNA synthetase in complex with asparagine-AMP | | Descriptor: | 4-AMINO-1,4-DIOXOBUTAN-2-AMINIUM ADENOSINE-5'-MONOPHOSPHATE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8H58
 
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
2VBE
 
 | | Tailspike protein of bacteriophage Sf6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Barbirz, S, Mueller, J.J, Freiberg, A, Heinemann, U, Seckler, R. | | Deposit date: | 2007-09-12 | | Release date: | 2008-04-22 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6.
Structure, 16, 2008
|
|
4WIZ
 
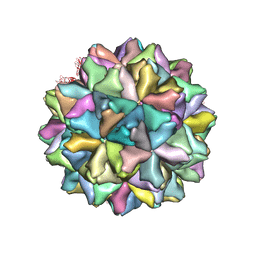 | | Crystal structure of Grouper nervous necrosis virus-like particle at 3.6A | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Chen, N.C, Chen, C.J, Yoshimura, M, Guan, H.H, Chen, T.Y. | | Deposit date: | 2014-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
Plos Pathog., 11, 2015
|
|
7ZWN
 
 | | Crystal structure of human BCL6 BTB domain in complex with a WVIP peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, B-cell lymphoma 6 protein, ... | | Authors: | Shetty, K, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
8H75
 
 | | FGFR2 in complex with YJ001 | | Descriptor: | CHLORIDE ION, Fibroblast growth factor receptor 2, Tinengotinib | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2022-10-19 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | FGFR2 in complex with YJ001
To Be Published
|
|
4WQB
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - bisulfite soak | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEME C, IODIDE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5013 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
2VC0
 
 | |
2V83
 
 | |
8H7Q
 
 | | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution
To Be Published
|
|
