7SWJ
 
 | | KirBac1.1 mutant - I131C | | Descriptor: | Inward rectifier potassium channel | | Authors: | Amani, R, Wylie, B.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Water Accessibility Refinement of the Extended Structure of KirBac1.1 in the Closed State.
Front Mol Biosci, 8, 2021
|
|
8CWX
 
 | |
1GJM
 
 | |
7X6O
 
 | |
6ZQM
 
 | | bovine ATP synthase monomer state 2 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RS6
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to GMPCPP-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2024
|
|
6MDN
 
 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MDM
 
 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6S8E
 
 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
7AI7
 
 | | MutS in Intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*TP*TP*AP*GP*CP*TP*TP*AP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*TP*AP*AP*CP*TP*AP*AP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AI5
 
 | | MutS in Scanning state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*AP*AP*TP*TP*CP*GP*CP*CP*CP*TP*AP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*TP*AP*GP*GP*GP*CP*GP*AP*AP*TP*TP*GP*GP*GP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Fernandez-Leiro, R, Bhairosing-Kok, D, Sixma, T.K, Lamers, M.H. | | Deposit date: | 2020-09-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The selection process of licensing a DNA mismatch for repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7SUN
 
 | |
7SJR
 
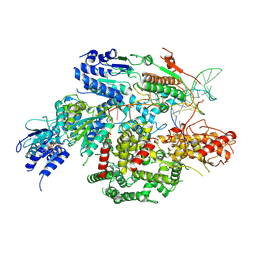 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | Descriptor: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | Authors: | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
4U4S
 
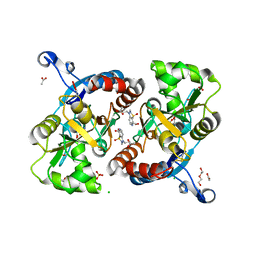 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM25 at 1.90 A resolution. | | Descriptor: | 4-ethyl-3,4-dihydro-2H-pyrido[4,3-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Noerholm, A.B, Deva, T, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
6OWS
 
 | |
1MPW
 
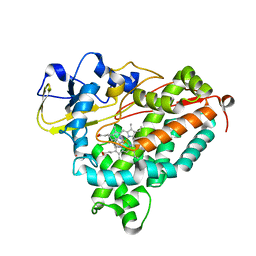 | | Molecular Recognition in (+)-a-Pinene Oxidation by Cytochrome P450cam | | Descriptor: | (+)-alpha-Pinene, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Bell, S.G, Chen, X, Sowden, R.J, Xu, F, Willams, J.N, Wong, L.-L, Rao, Z. | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam
J.Am.Chem.Soc., 125, 2003
|
|
5XOJ
 
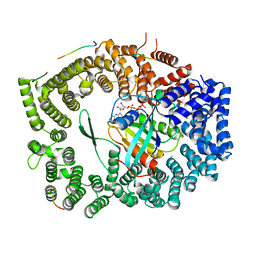 | | Crystal structure of Xpo1p-PKI-Nup42p-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p
Genes Cells, 22, 2017
|
|
7TYL
 
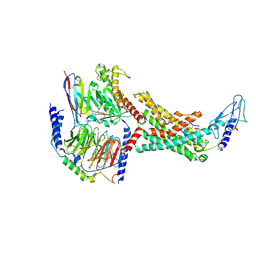 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYO
 
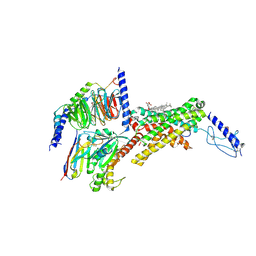 | | Calcitonin receptor in complex with Gs and human calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYN
 
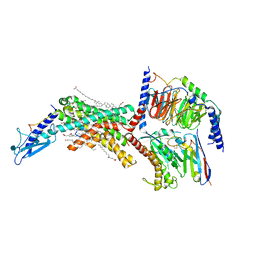 | | Calcitonin Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYI
 
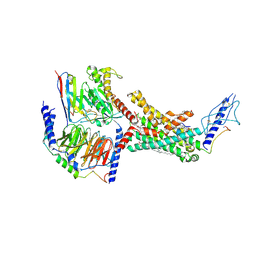 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, CT-like state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
8V6V
 
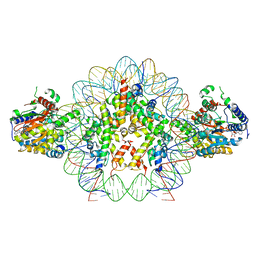 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
6USF
 
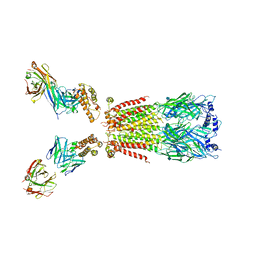 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
1MXU
 
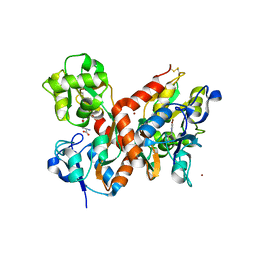 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) in complex with bromo-willardiine (Control for the crystal titration experiments) | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
