6GLD
 
 | |
6JDE
 
 | | crystal structure of a DNA repair protein | | Descriptor: | Putative DNA repair helicase RadD, ZINC ION | | Authors: | Yan, X.X, Tang, Q. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel ATPase RadD from Escherichia coli.
Proteins, 87, 2019
|
|
2VL7
 
 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
4BE7
 
 | | MUTANT (K220R) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
5CV2
 
 | |
3I3Q
 
 | | Crystal Structure of AlkB in complex with Mn(II) and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB, MANGANESE (II) ION | | Authors: | Yu, B, Hunt, J.F. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ZBB
 
 | | Structure of the 4_601_167 Tetranucleosome | | Descriptor: | DNA STRAND 1 (ARBITRARY MODEL SEQUENCE), DNA STRAND 2 (ARBITRARY MODEL SEQUENCE), HISTONE H3, ... | | Authors: | Schalch, T, Duda, S, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | X-ray structure of a tetranucleosome and its implications for the chromatin fibre.
Nature, 436, 2005
|
|
4RGH
 
 | |
1D4B
 
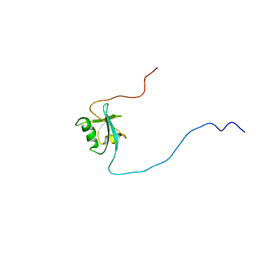 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
2Y3M
 
 | | Structure of the extra-membranous domain of the secretin HofQ from Actinobacillus actinomycetemcomitans | | Descriptor: | FORMIC ACID, GLYCEROL, PROTEIN TRANSPORT PROTEIN HOFQ | | Authors: | Tarry, M, Jaaskelainen, M, Tuominen, H, Ihalin, R, Hogbom, M. | | Deposit date: | 2010-12-21 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Extra-Membranous Domains of the Competence Protein Hofq Show DNA Binding, Flexibility and a Shared Fold with Type I Kh Domains.
J.Mol.Biol., 409, 2011
|
|
5BOC
 
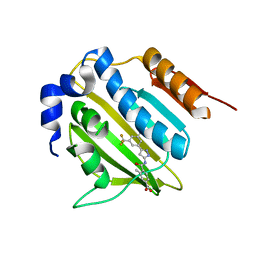 | | Crystal structure of topoisomerase ParE inhibitor | | Descriptor: | 3-methyl-4-({3-[3-methyl-5-(trifluoromethyl)phenyl]-1H-pyrazol-5-yl}carbamoyl)benzoic acid, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G.Y, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
To be published
|
|
5BOD
 
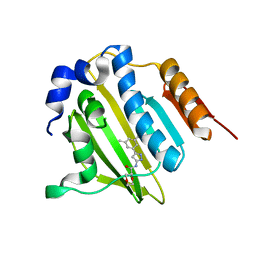 | | Crystal structure of Streptococcus pneumonia ParE inhibitor | | Descriptor: | (2R)-N-[3-(3,5-dimethylphenyl)-1H-pyrazol-5-yl]-1,4-dioxane-2-carboxamide, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
to be published
|
|
1XCI
 
 | | Mispair Aligned N3T-Butyl-N3T Interstrand Crosslink | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*(TTM)P*TP*TP*TP*CP*G)-3' | | Authors: | da Silva, M.W, Bierbryer, R.G, Wilds, C.J, Noronha, A.M, Colvin, O.M, Miller, P.S, Gamcsik, M.P. | | Deposit date: | 2004-09-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrastrand base-stacking buttresses widening of major groove in interstrand cross-linked B-DNA.
Bioorg.Med.Chem., 13, 2005
|
|
1JUA
 
 | |
4BEC
 
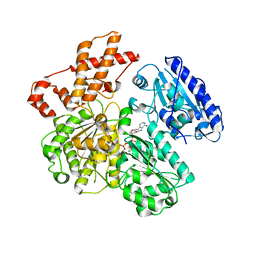 | | MUTANT (K220A) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BEB
 
 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
1MUY
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI | | Descriptor: | ADENINE GLYCOSYLASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
1FUF
 
 | | CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE | | Descriptor: | (5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2000-09-15 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
2KJN
 
 | |
1Z5Z
 
 | | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain | | Descriptor: | Helicase of the snf2/rad54 family | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA
Cell(Cambridge,Mass.), 121, 2005
|
|
2KJO
 
 | |
4E8U
 
 | |
2WKD
 
 | |
1MUN
 
 | |
2JUH
 
 | |
