6RZM
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:5(Iodine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(3-iodanylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
4A2B
 
 | |
6GHB
 
 | |
2PZX
 
 | | Structure of the methuselah ectodomain with peptide inhibitor | | Descriptor: | G-protein coupled receptor Mth | | Authors: | West Jr, A.P, Ja, W.W, Delker, S.L, Bjorkman, P.J, Benzer, S, Roberts, R.W. | | Deposit date: | 2007-05-18 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor.
Nat.Chem.Biol., 3, 2007
|
|
7EME
 
 | | Putative Leptospira interrogans recombinant L-amino acid oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase | | Authors: | Vaigundan, D, Yuvaraj, I, Krishnaswamy, P.R, Sekar, K, Murthy, M.R.N, Sunita, P. | | Deposit date: | 2021-04-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural characterization of a putative recombinant L-amino acid oxidase from Leptospira interrogans
Curr.Sci., 123, 2022
|
|
3PG9
 
 | | Thermotoga maritima DAH7P synthase in complex with inhibitor | | Descriptor: | AZIDE ION, CHLORIDE ION, NITRATE ION, ... | | Authors: | Cross, P.J, Dobson, R.C.J, Patchett, M.L, Parker, E.J. | | Deposit date: | 2010-10-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tyrosine latching of a regulatory gate affords allosteric control of aromatic amino acid biosynthesis
J.Biol.Chem., 286, 2011
|
|
5AWM
 
 | |
7RUJ
 
 | | E. coli cysteine desulfurase SufS N99A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
7RW3
 
 | | E. coli cysteine desulfurase SufS N99D | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
6P3K
 
 | | Crystal structure of LigU(C100S) | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6P3H
 
 | | Crystal structure of LigU(K66M) bound to substrate | | Descriptor: | (1E)-4-oxobut-1-ene-1,2,4-tricarboxylic acid, (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
6P3J
 
 | | Crystal structure of LigU | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
7DB0
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB2
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in GS-dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB4
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP012- and glutathione-bound form | | Descriptor: | 1-[(4-fluorophenyl)methyl]-2,2-bis(oxidanylidene)thieno[3,2-c][1,2]thiazin-4-one, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB1
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in dimedone- and glutathione-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAX
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-bound form | | Descriptor: | 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7000097 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAZ
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP015- and GSH-bound form | | Descriptor: | 6-(6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl)pyridin-3-amine, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB3
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP011-bound form | | Descriptor: | 4-bromanyl-2-[[2-[(E)-1-(3-methoxyphenyl)ethylideneamino]propan-2-ylamino]methyl]phenol, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAY
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-, and GSH-bound form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
5N6T
 
 | |
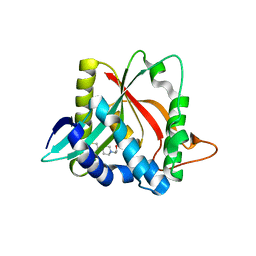
3V8I
 
 | |
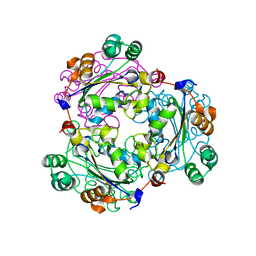
3WX8
 
 | |
2ORW
 
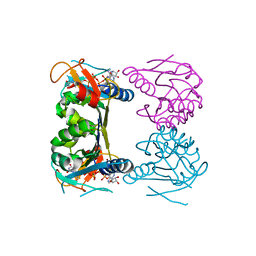 | | Thermotoga maritima thymidine kinase 1 like enzyme in complex with TP4A | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P4-(5'-(2'-DEOXY-THYMIDYL))TETRAPHOSPHATE, Thymidine kinase, ... | | Authors: | Segura-Pena, D, Lutz, S, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2007-02-04 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of ATP to TK1-like Enzymes Is Associated with a Conformational Change in the Quaternary Structure.
J.Mol.Biol., 369, 2007
|
|
5YXW
 
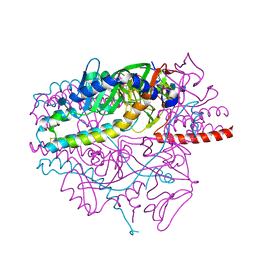 | | Crystal structure of the prefusion form of measles virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
