2HOY
 
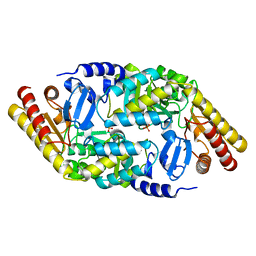 | | Inter-subunit signaling in GSAM | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) apo-form, PHOSPHATE ION | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NAY
 
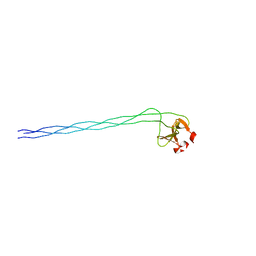 | | GPP-Foldon:X-ray structure | | Descriptor: | Collagen-like peptide | | Authors: | Stetefeld, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Collagen Stabilization at Atomic Level. Crystal Structure of Designed (GlyProPro)(10)foldon
Structure, 11, 2003
|
|
3V2R
 
 | | COMPcc in complex with fatty acids | | Descriptor: | Cartilage Oligomerization matrix protein (coiled-coil domain), OLEIC ACID | | Authors: | Stetefeld, J. | | Deposit date: | 2011-12-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | COMPcc in complex with fatty acids
To be Published
|
|
1PXU
 
 | |
1PZ8
 
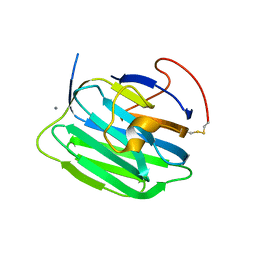 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ9
 
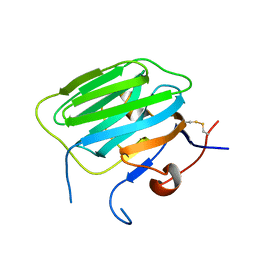 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ7
 
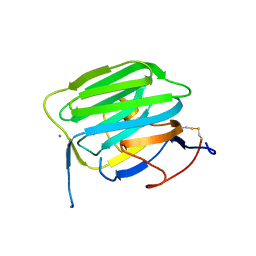 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1Q56
 
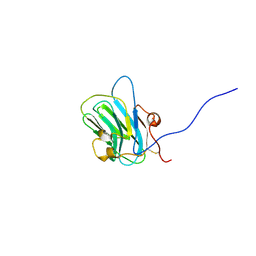 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1PFX
 
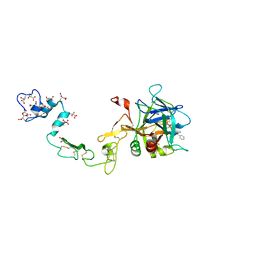 | | PORCINE FACTOR IXA | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, FACTOR IXA | | Authors: | Brandstetter, H, Bauer, M, Huber, R, Lollar, P, Bode, W. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1FAX
 
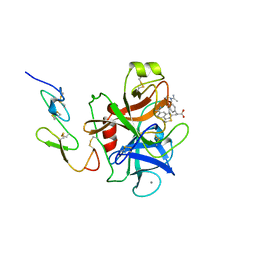 | | COAGULATION FACTOR XA INHIBITOR COMPLEX | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, FACTOR XA | | Authors: | Brandstetter, H, Engh, R.A. | | Deposit date: | 1996-08-23 | | Release date: | 1997-10-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of active site-inhibited clotting factor Xa. Implications for drug design and substrate recognition.
J.Biol.Chem., 271, 1996
|
|
1JC7
 
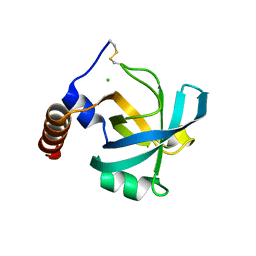 | |
1JJ9
 
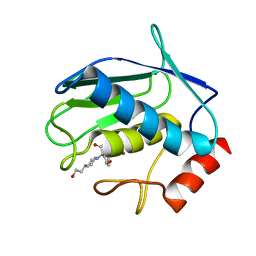 | | Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition | | Descriptor: | 2-HYDROXY-5-[4-(2-HYDROXY-ETHYL)-PIPERIDIN-1-YL]-5-PHENYL-1H-PYRIMIDINE-4,6-DIONE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Brandstetter, H, Grams, F, Glitz, D, Lang, A, Huber, R, Bode, W, Krell, H.-W, Engh, R.A. | | Deposit date: | 2001-07-04 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8-A crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition.
J.Biol.Chem., 276, 2001
|
|
1JB3
 
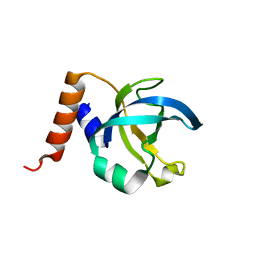 | |
1K32
 
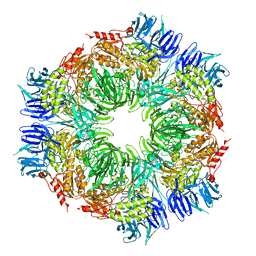 | |
1Y0Y
 
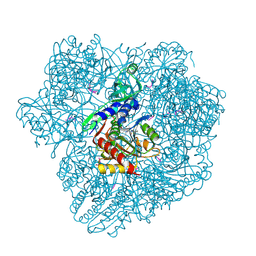 | |
1Y0R
 
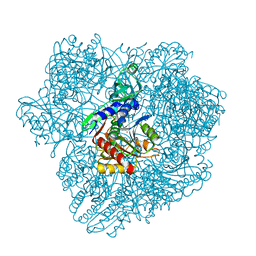 | |
8HMA
 
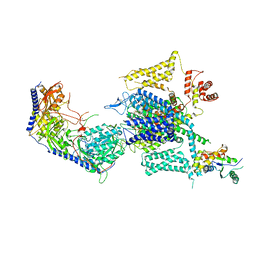 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8ER1
 
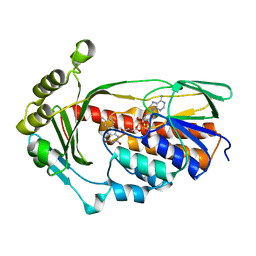 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
7EPW
 
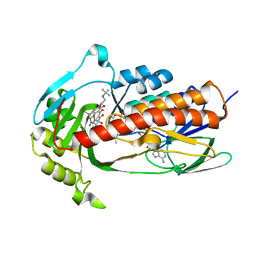 | | Crystal structure of monooxygenase Tet(X4) with tigecycline | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, TIGECYCLINE | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
7EPV
 
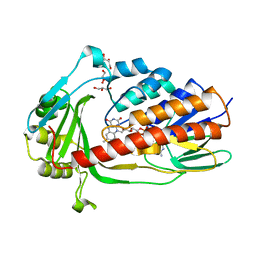 | | Crystal structure of tigecycline degrading monooxygenase Tet(X4) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, GLYCEROL | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
4Y0P
 
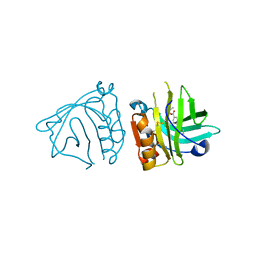 | | Bovine beta-lactoglobulin complex with tetracaine (BLG-TET) | | Descriptor: | Beta-lactoglobulin, Tetracaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
1M5B
 
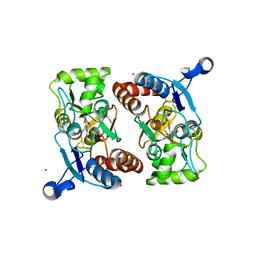 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
2WZN
 
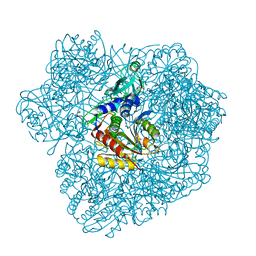 | | 3d structure of TET3 from Pyrococcus horikoshii | | Descriptor: | 354AA LONG HYPOTHETICAL OPERON PROTEIN FRV, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rosenbaum, E, Dura, M.A, Vellieux, F.M, Franzetti, B. | | Deposit date: | 2009-12-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural and Biochemical Characterizations of a Novel Tet Peptidase Complex from Pyrococcus Horikoshii Reveal an Integrated Peptide Degradation System in Hyperthermophilic Archaea.
Mol.Microbiol., 72, 2009
|
|
4V4E
 
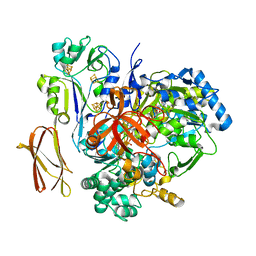 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
5OXF
 
 | |
