2NWU
 
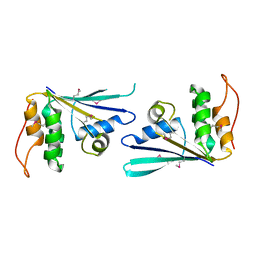 | |
7MJZ
 
 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
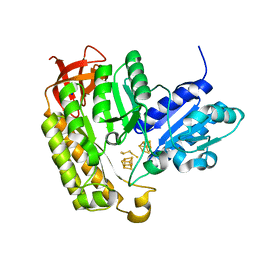
2XI3
 
 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
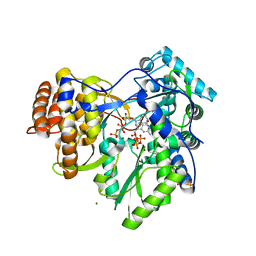
2XHV
 
 | | HCV-J4 NS5B Polymerase Point Mutant Orthorhombic Crystal Form | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
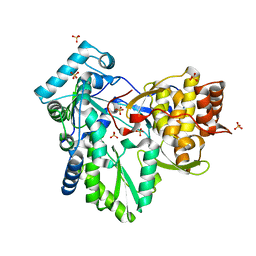
3KWR
 
 | |
2PZZ
 
 | |
1FSI
 
 | | CRYSTAL STRUCTURE OF CYCLIC NUCLEOTIDE PHOSPHODIESTERASE OF APPR>P FROM ARABIDOPSIS THALIANA | | Descriptor: | CYCLIC PHOSPHODIESTERASE, SULFATE ION | | Authors: | Hofmann, A, Zdanov, A, Genschik, P, Filipowicz, W, Ruvinov, S, Wlodawer, A. | | Deposit date: | 2000-09-10 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction.
EMBO J., 19, 2000
|
|
2FYM
 
 | |
3L29
 
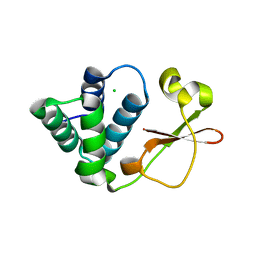 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | Descriptor: | CHLORIDE ION, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
1X48
 
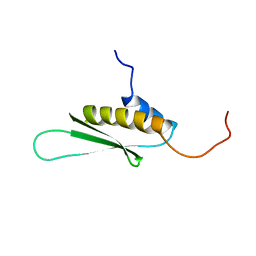 | | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Tarada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1X49
 
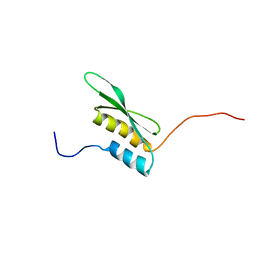 | | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
3V97
 
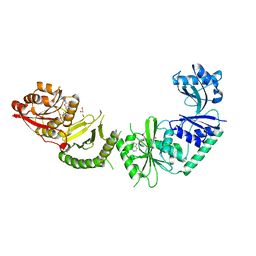 | |
6GV4
 
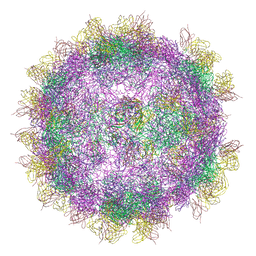 | | High-resolution Cryo-EM of Fab-labeled human parechovirus 3 | | Descriptor: | AT12-015 antibody variable heavy, AT12-015 antibody variable light, RNA (5'-R(*UP*GP*GP*UP*AP*UP*UP*U)-3'), ... | | Authors: | Domanska, A, Flatt, J.W, Jukonen, J.J.J, Geraets, J.A, Butcher, S.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.
J.Virol., 93, 2019
|
|
3ERC
 
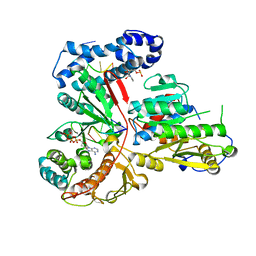 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase with three fragments of RNA and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3V8V
 
 | |
2J04
 
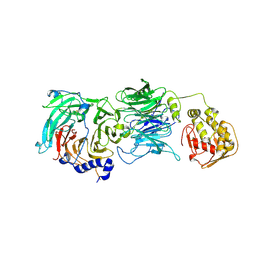 | | The tau60-tau91 subcomplex of yeast transcription factor IIIC | | Descriptor: | HYPOTHETICAL PROTEIN YPL007C, YDR362CP | | Authors: | Mylona, A, Fernandez-Tornero, C, Legrand, P, Muller, C.W. | | Deposit date: | 2006-07-31 | | Release date: | 2006-10-23 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Tau60/Deltatau91 Subcomplex of Yeast Transcription Factor Iiic: Insights Into Preinitiation Complex Assembly
Mol.Cell, 24, 2006
|
|
6RKQ
 
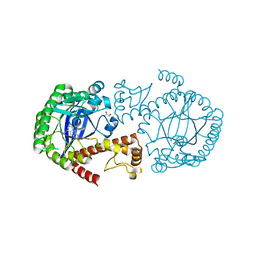 | | Crystal Structure of TGT in complex with N2-methyl-8-(prop-1-yn-1-yl)-3H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | (8~{R})-~{N}2-methyl-8-prop-1-ynyl-7,8-dihydro-3~{H}-imidazo[4,5-g]quinazoline-2,6-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
6RKT
 
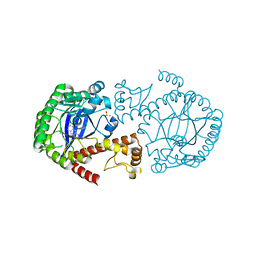 | | Crystal Structure of TGT in complex with N2-methyl-1H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
4HDH
 
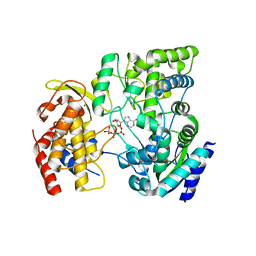 | | Crystal Structure of viral RdRp in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polyprotein, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
8JYI
 
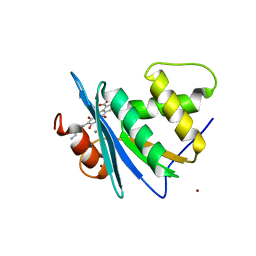 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
2EGW
 
 | |
2E7E
 
 | |
2JX2
 
 | | Solution conformation of RNA-bound NELF-E RRM | | Descriptor: | Negative elongation factor E | | Authors: | Jampani, N, Schweimer, K, Wenzel, S, Woehrl, B.M, Roesch, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NELF-E RRM Undergoes Major Structural Changes in Flexible Protein Regions on Target RNA Binding
Biochemistry, 47, 2008
|
|
1MNM
 
 | |
3J1O
 
 | |
