9FE6
 
 | | Short-chain dehydrogenase/reductase (SDR) from Thermus caliditerrae | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Kapur, B, Nar, H. | | Deposit date: | 2024-05-17 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | In silico enzyme screening identifies an SDR ketoreductase from Thermus caliditerrae as an attractive biocatalyst and promising candidate for protein engineering
Front Chem Biol, 2024
|
|
6X64
 
 | | Legionella pneumophila Dot T4SS PR | | Descriptor: | Type IV secretion system unknown protein fragment | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
5HFO
 
 | | CRYSTAL STRUCTURE OF OXA-232 BETA-LACTAMASE | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Retailleau, P, Oueslati, S, Cisse, C, Nordmann, P, Naas, T, Iorga, B. | | Deposit date: | 2016-01-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Role of Arginine 214 in the Substrate Specificity of OXA-48.
Antimicrob.Agents Chemother., 64, 2020
|
|
6XBQ
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase in complex with carboxy-carba(dethia)-CoA | | Descriptor: | CARBOXYMETHYLDETHIA COENZYME *A, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase in complex with carboxy-carba(dethia)-CoA
To Be Published
|
|
6XBT
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase (Q60A) in complex with 2-nitronate-propionyl-CoA | | Descriptor: | COBALT (II) ION, COENZYME A, Methylmalonyl-CoA epimerase, ... | | Authors: | Stunkard, L.M, Boram, T.J, Benjamin, A.B, Bower, J.B, Lohman, J.R. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Streptomyces coelicolor methylmalonyl-CoA epimerase (Q60A) in complex with 2-nitronate-propionyl-CoA
To Be Published
|
|
9CIA
 
 | | T cell receptor complex | | Descriptor: | CHOLESTEROL, T cell receptor delta constant, T cell receptor gamma constant 1, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
9FSB
 
 | | Coxsackievirus B3 3C protease in P121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Coxsackievirus B3 3C protease in P121 spacegroup
To Be Published
|
|
9CIL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T41V/S59A at cryogenic temperature
To Be Published
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
9CII
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, Robinson, A.C, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36R at cryogenic temperature
To Be Published
|
|
7N06
 
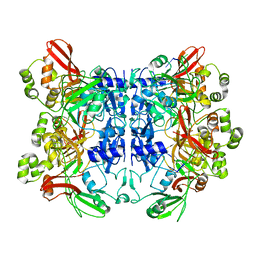 | | SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state | | Descriptor: | RNA (5'-R(*AP*UP*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
8VR6
 
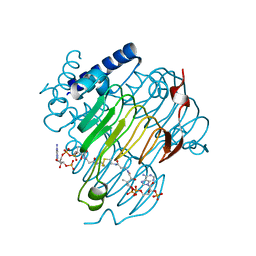 | | crystal structure of the Pcryo_0619 N-acetryltransferase from Psychrobacter cryohalolentis K5 in the presence of CoA-disulfide | | Descriptor: | Acetyltransferase, putative, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
9CIJ
 
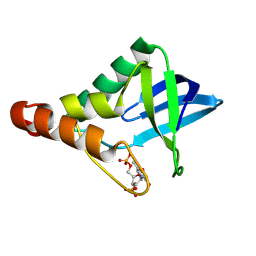 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, L.J, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature
To Be Published
|
|
9EOU
 
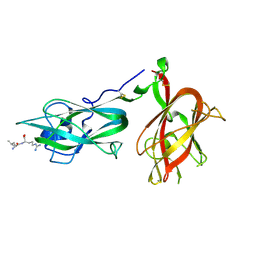 | |
9CIK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Zhang, Y, Schlessman, J.L, SIegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-07-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature
To Be Published
|
|
9CI8
 
 | | T cell receptor complex | | Descriptor: | T cell receptor delta constant, T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
9ENP
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
9EV6
 
 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO), C-terminal truncated construct | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 2024
|
|
9BCG
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 2024
|
|
8YLC
 
 | | The crystal structure of PDE4D with Amentoflavone | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, 8-[5-[5,7-bis(oxidanyl)-4-oxidanylidene-chromen-2-yl]-2-oxidanyl-phenyl]-2-(4-hydroxyphenyl)-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.30003715 Å) | | Cite: | Discovery of amentoflavone as a natural PDE4 inhibitor with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8P5P
 
 | | Structure of TECPR1 N-terminal DysF domain | | Descriptor: | GLYCEROL, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Boyle, K.B, Elliott, P.R, Randow, F. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TECPR1 conjugates LC3 to damaged endomembranes upon detection of sphingomyelin exposure.
Embo J., 42, 2023
|
|
9G11
 
 | |
9ENQ
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
7N33
 
 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
6XP4
 
 | |
