8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G9L
 
 | | DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
7U66
 
 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxyguanosinetriphosphate triphosphohydrolase, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Dillard, L.B, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GN6
 
 | | Crystallization of Sialidase from Porphyromonas gingivalis | | Descriptor: | Sialidase, UNKNOWN LIGAND | | Authors: | Dong, W.B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and enzymatic characterization of the sialidase SiaPG from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G9N
 
 | | Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G69
 
 | | Wildtype PTP1b in complex with DES5743 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(5-ethyl-3-methyl-1H-pyrazol-1-yl)aniline, MAGNESIUM ION, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G67
 
 | | Wildtype PTP1b in complex with DES4884 | | Descriptor: | 6-methyl-4-(piperazin-1-yl)-2-(trifluoromethyl)quinoline, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
1ADI
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE AT PH 6.5 AND 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
8QEV
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
8QUG
 
 | | KRAS-G12C in Complex with Compound 1 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[2-[(2S)-2-methyl-1,4-diazepan-1-yl]pyrimidin-4-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fischer, G, Kratochvil, B. | | Deposit date: | 2023-10-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Targeting cancer with small molecule pan-KRAS degraders
Biorxiv, 2023
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
6T1G
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
8G6A
 
 | | Wildtype PTP1b in complex with DES6016 | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8GS9
 
 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | Descriptor: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | Authors: | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8G25
 
 | |
6NLI
 
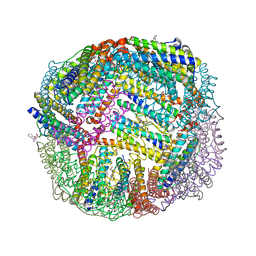 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 11) | | Descriptor: | 4-{[(2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
7QBG
 
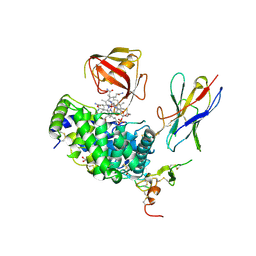 | | TC:CD320 in complex with nanobody TC-Nb4 | | Descriptor: | Anti-TC:CD320 nanobody TC-Nb4, CALCIUM ION, CD320 antigen, ... | | Authors: | Bloch, J.S, Locher, K.P. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Faseb J., 36, 2022
|
|
6NKP
 
 | |
8QC8
 
 | |
8G68
 
 | | Wildtype PTP1b in complex with DES5742 | | Descriptor: | 4-(3-ethyl-5-methyl-1H-pyrazol-1-yl)aniline, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G65
 
 | | Wildtype PTP1b in complex with DES4799 | | Descriptor: | 4-(3,5-dimethyl-1H-pyrazol-1-yl)aniline, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8QC2
 
 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
