3EL3
 
 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
3F6U
 
 | | Crystal structure of human Activated Protein C (APC) complexed with PPACK | | Descriptor: | CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Schmidt, A.E, Padmanabhan, K, Underwood, M.C, Bode, W, Mather, T, Bajaj, S.P. | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC).
J.Biol.Chem., 277, 2002
|
|
6JEM
 
 | | Structure of Phytolacca americana UGT2 complexed with UDP-2fluoro-glucose and resveratrol | | Descriptor: | Glycosyltransferase, RESVERATROL, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
6JGE
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4B7R
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
6JGS
 
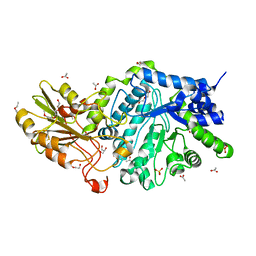 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2OYU
 
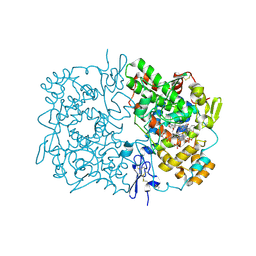 | | Indomethacin-(S)-alpha-ethyl-ethanolamide bound to Cyclooxygenase-1 | | Descriptor: | 2-[1-(4-CHLOROBENZOYL)-5-METHOXY-2-METHYL-1H-INDOL-3-YL]-N-[(1S)-1-(HYDROXYMETHYL)PROPYL]ACETAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Harman, C.A, Garavito, R.M. | | Deposit date: | 2007-02-23 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-alpha-substituted indomethacin ethanolamides.
J.Biol.Chem., 282, 2007
|
|
6KOB
 
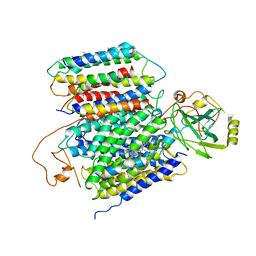 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1YR1
 
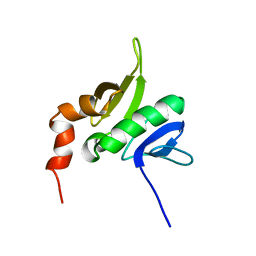 | |
6LC5
 
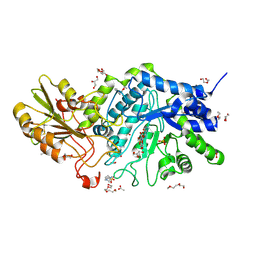 | | Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-17 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
1M5C
 
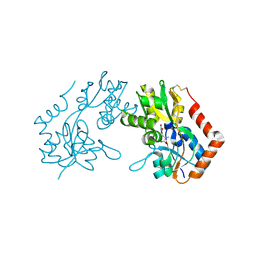 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1UEX
 
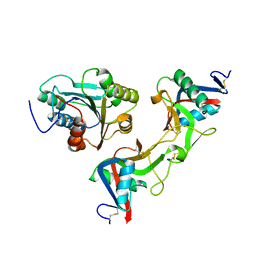 | | Crystal structure of von Willebrand Factor A1 domain complexed with snake venom bitiscetin | | Descriptor: | bitiscetin alpha chain, bitiscetin beta chain, von Willebrand Factor | | Authors: | Maita, N, Nishio, K, Nishimoto, E, Matsui, T, Shikamoto, Y, Morita, T, Sadler, J.E, Mizuno, H. | | Deposit date: | 2003-05-22 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin. Insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins.
J.Biol.Chem., 278, 2003
|
|
1H9Y
 
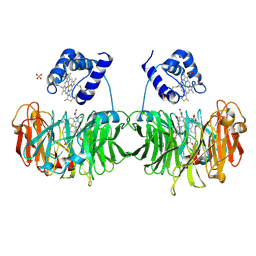 | |
4ND3
 
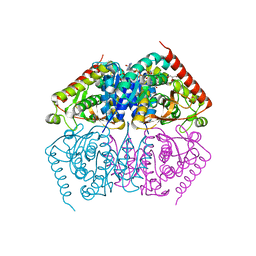 | |
4NYY
 
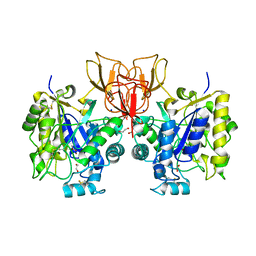 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1WWN
 
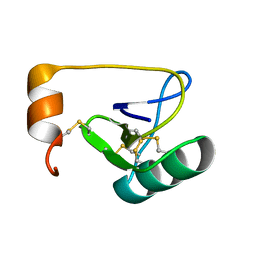 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
4ND4
 
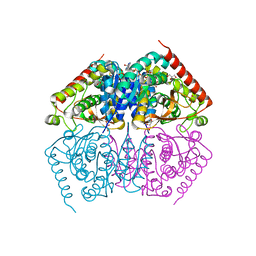 | |
7XWT
 
 | |
7XSW
 
 | | Structure of SARS-CoV-2 antibody S309 with GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Lambda Chain, ... | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-05-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7XWV
 
 | |
7XWC
 
 | |
1H9H
 
 | | COMPLEX OF EETI-II WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7Y9P
 
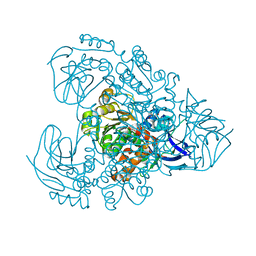 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
4NZ1
 
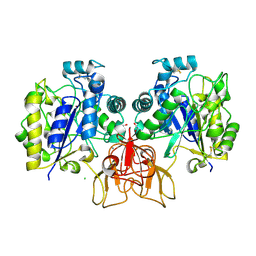 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1H9I
 
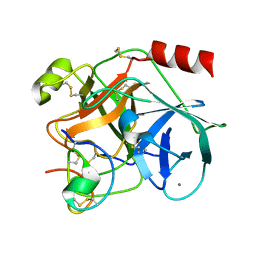 | | COMPLEX OF EETI-II MUTANT WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
