124D
 
 | |
133L
 
 | |
134L
 
 | |
135L
 
 | |
1IFH
 
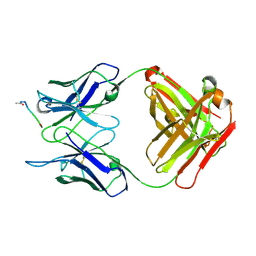 | |
1HEM
 
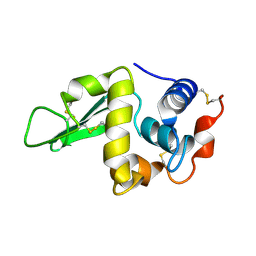 | |
1HER
 
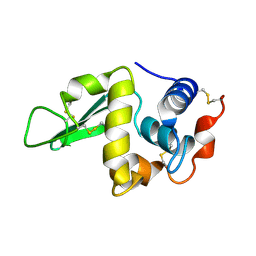 | |
5CCP
 
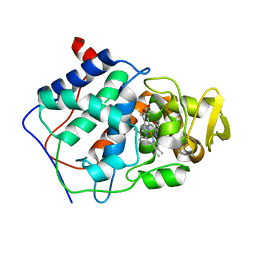 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
2LAL
 
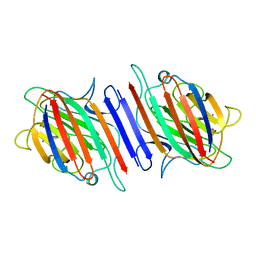 | | CRYSTAL STRUCTURE DETERMINATION AND REFINEMENT AT 2.3 ANGSTROMS RESOLUTION OF THE LENTIL LECTIN | | Descriptor: | CALCIUM ION, LENTIL LECTIN (ALPHA CHAIN), LENTIL LECTIN (BETA CHAIN), ... | | Authors: | Loris, R, Steyaert, J, Maes, D, Lisgarten, J, Pickersgill, R, Wyns, L. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
3CD4
 
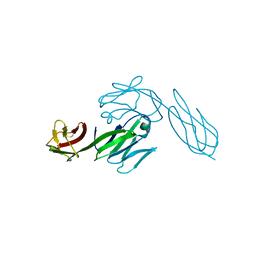 | |
3CI2
 
 | |
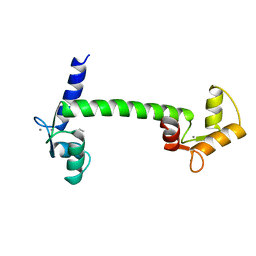
1CLL
 
 | |
1HHG
 
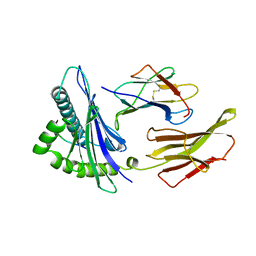 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), HIV-1 GP120 ENVELOPE PROTEIN (RESIDUES 195-207) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
1COR
 
 | |
104L
 
 | |
103L
 
 | |
1AAN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
1NDK
 
 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
2HOA
 
 | | STRUCTURE DETERMINATION OF THE ANTP(C39->S) HOMEODOMAIN FROM NUCLEAR MAGNETIC RESONANCE DATA IN SOLUTION USING A NOVEL STRATEGY FOR THE STRUCTURE CALCULATION WITH THE PROGRAMS DIANA, CALIBA, HABAS AND GLOMSA | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Guntert, P, Qian, Y.-Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1992-04-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA.
J.Mol.Biol., 217, 1991
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
1SDA
 
 | | CRYSTAL STRUCTURE OF PEROXYNITRITE-MODIFIED BOVINE CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Smith, C.D, Carson, M, Van Der Woerd, M, Chen, J, Ischiropoulos, H, Beckman, J.S. | | Deposit date: | 1993-01-13 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of peroxynitrite-modified bovine Cu,Zn superoxide dismutase.
Arch.Biochem.Biophys., 299, 1992
|
|
1CMB
 
 | |
1HEO
 
 | |
1ALB
 
 | |
