6JEL
 
 | | Structure of Phytolacca americana apo UGT2 | | Descriptor: | Glycosyltransferase | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
6JKU
 
 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | Authors: | Manjunath, L, Bose, S, Subramanian, R. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
1E20
 
 | | The FMN binding protein AtHal3 | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, HALOTOLERANCE PROTEIN HAL3, ... | | Authors: | Albert, A, Martinez-Ripoll, M, Espinosa-Ruiz, A, Yenush, L, Culianez-Macia, F.A, Serrano, R. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-11 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The X-Ray Structure of the Fmn-Binding Protein Athal3 Provides the Structural Basis for the Activity of a Regulatory Subunit Involved in Signal Transduction
Structure, 8, 2000
|
|
8U8V
 
 | | Cryo-EM structure of Substrate ATP Bound in the Insertion Site (IS) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
8U8U
 
 | | Cryo-EM Structure of Cognate Substrate ATP Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
3LU6
 
 | | Human serum albumin in complex with compound 1 | | Descriptor: | Serum albumin, [(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]acetic acid | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
8UPK
 
 | |
3LU7
 
 | | Human serum albumin in complex with compound 2 | | Descriptor: | 4-[(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]butanoic acid, PHOSPHATE ION, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
8V5T
 
 | | Crystal structure of Alzheimers disease phospholipase D3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, GLYCEROL, ... | | Authors: | Ishii, K, Hermans, S.J, Nero, T.L, Gorman, M.A, Parker, M.W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Alzheimer's disease phospholipase D3 provides a molecular basis for understanding its normal and pathological functions.
Febs J., 2024
|
|
8H5A
 
 | |
8H58
 
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
3CTY
 
 | |
3L91
 
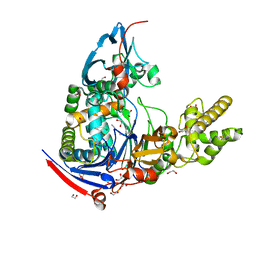 | | Structure of Pseudomonas aerugionsa PvdQ bound to octanoate | | Descriptor: | 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase pvdQ subunit alpha, Acyl-homoserine lactone acylase pvdQ subunit beta, ... | | Authors: | Drake, E.J, Gulick, A.M. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis.
Acs Chem.Biol., 6, 2011
|
|
6H8J
 
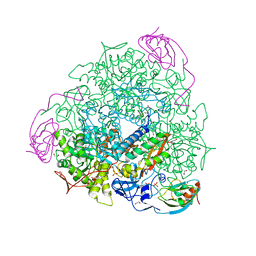 | | 1.45 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPTO | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-08-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into Urease Inhibition by N-( n-Butyl) Phosphoric Triamide through an Integrated Structural and Kinetic Approach.
J.Agric.Food Chem., 67, 2019
|
|
3LQR
 
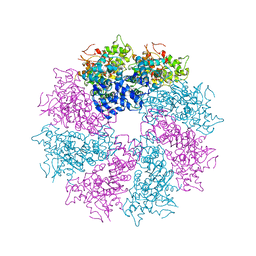 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
3DHB
 
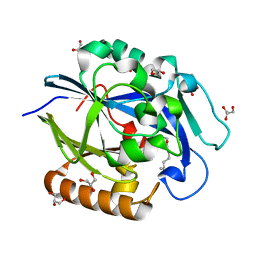 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
8I2E
 
 | |
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI2
 
 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8I2F
 
 | |
8I2D
 
 | | Crystal structure of Bacillus subtilis LytE | | Descriptor: | Probable peptidoglycan endopeptidase LytE | | Authors: | Tandukar, S, Kwon, E, Kim, D.Y. | | Deposit date: | 2023-01-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural insights into the regulation of peptidoglycan DL-endopeptidases by inhibitory protein IseA.
Structure, 31, 2023
|
|
3DWV
 
 | |
