4QNW
 
 | |
4HEQ
 
 | | The crystal structure of flavodoxin from Desulfovibrio gigas | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Dimeric Flavodoxin from Desulfovibrio gigas Suggests a Potential Binding Region for the Electron-Transferring Partner
Int J Mol Sci, 14, 2013
|
|
3L0S
 
 | | Crystal structures of Zinc, Cobalt and Iron containing Adenylate kinase from Gram-negative bacteria Desulfovibrio gigas | | Descriptor: | Adenylate kinase, COBALT (II) ION, D(-)-TARTARIC ACID | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
2PAN
 
 | | Crystal structure of E. coli glyoxylate carboligase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, Glyoxylate carboligase, ... | | Authors: | Kaplun, A, Chipman, D.M, Barak, Z, Vyazmensky, M, Shaanan, B. | | Deposit date: | 2007-03-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes.
Nat.Chem.Biol., 4, 2008
|
|
3UTH
 
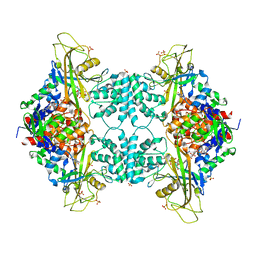 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with substrate UDP-Galp in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
1DXG
 
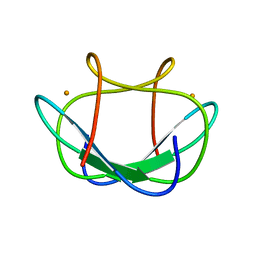 | |
3UKK
 
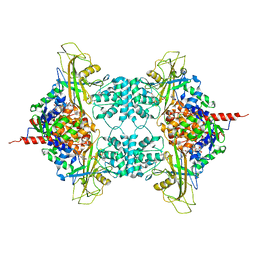 | |
3UKQ
 
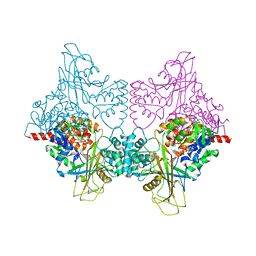 | |
1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
3UTG
 
 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with UDP in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3UTF
 
 | |
3UKL
 
 | |
3L0P
 
 | | Crystal structures of Iron containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | Adenylate kinase, FE (III) ION, GLYCEROL | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
5AF7
 
 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BMA
 
 | | structural of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase | | Descriptor: | GLYCEROL, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, HurtadoGuerrero, R, vanAalten, D.M.F. | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Genetic and Structural Validation of Aspergillus Fumigatus Udp-N-Acetylglucosamine Pyrophosphorylase as an Antifungal Target.
Mol.Microbiol., 89, 2013
|
|
3UTE
 
 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase sulfate complex | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3UKA
 
 | |
3QGH
 
 | |
1RDG
 
 | |
3QGF
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(6-chloropyridazin-3-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGI
 
 | |
3QGG
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, N-cyclopropyl-6-[(3R)-3-{[4-(trifluoromethoxy)benzyl]carbamoyl}-4-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazin-1-yl]pyridazine-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QGD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5UBM
 
 | | Crystal structure of human C1s in complex with inhibitor gigastasin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C1s subcomponent, Gigastasin | | Authors: | Pang, S.S, Whisstock, J.C. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Basis for Complement Inhibition by Gigastasin, a Protease Inhibitor from the Giant Amazon Leech.
J. Immunol., 199, 2017
|
|
5TUP
 
 | | X-ray Crystal Structure of the Aspergillus fumigatus Sliding Clamp | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Bruning, J.B, Marshall, A.C, Kroker, A.J, Wegener, K.L, Rajapaksha, H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with Human p21.
FEBS J., 284, 2017
|
|
