2MNU
 
 | |
2MMY
 
 | |
2H6X
 
 | |
2H7Q
 
 | | Cytochrome P450cam complexed with imidazole | | Descriptor: | Cytochrome P450-cam, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
2H8D
 
 | | Crystal structure of deoxy hemoglobin from Trematomus bernacchii at pH 8.4 | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, POTASSIUM ION, ... | | Authors: | Mazzarella, L, Vergara, A, Vitagliano, L, Merlino, A, Bonomi, G, Scala, S, Verde, C, di Prisco, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | High resolution crystal structure of deoxy hemoglobin from Trematomus bernacchii at different pH values: The role of histidine residues in modulating the strength of the root effect.
Proteins, 65, 2006
|
|
2MJN
 
 | |
2JYQ
 
 | | NMR structure of the apo v-Src SH2 domain | | Descriptor: | Tyrosine-protein kinase transforming protein Src | | Authors: | Taylor, J.D, Ababou, A, Williams, M.A, Ladbury, J.E. | | Deposit date: | 2007-12-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: Implications for drug design
Proteins, 73, 2008
|
|
2K1X
 
 | | NMR solution structure of M-crystallin in calcium free form (apo). | | Descriptor: | Beta/gama crystallin family protein | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2MEF
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-04 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEG
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS. | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2LWI
 
 | | Solution structure of H-RasT35S mutant protein in complex with Kobe2601 | | Descriptor: | 2-(2,4-dinitrophenyl)-N-(4-fluorophenyl)hydrazinecarbothioamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Araki, M, Tamura, A, Shima, F, Kataoka, T. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M1D
 
 | |
2M8H
 
 | |
2JYO
 
 | | NMR Solution structure of Human MIP-3alpha/CCL20 | | Descriptor: | C-C motif chemokine 20 (Small-inducible cytokine A20) (Macrophage inflammatory protein 3 alpha) (MIP-3-alpha) (Liver and activation-regulated chemokine) (CC chemokine LARC) (Beta chemokine exodus-1) | | Authors: | Chan, D.I, Hunter, H.N, Tack, B.F, Vogel, H.J. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Human macrophage inflammatory protein 3alpha: protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties.
Antimicrob.Agents Chemother., 52, 2008
|
|
2MGB
 
 | |
2JVI
 
 | |
2JRL
 
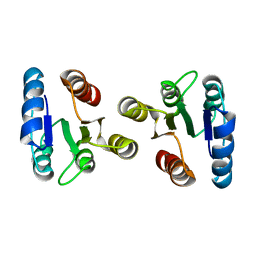 | | Solution structure of the beryllofluoride-activated NtrC4 receiver domain dimer | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Lee, C, Hong, E, Doucleff, M, Pelton, J.G, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Beryllofluoride-Activated NtrC4 Receiver Domain Dimer.
To be Published
|
|
2JVZ
 
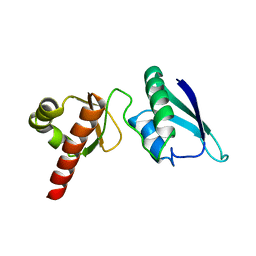 | | Solution NMR Structure of the Second and Third KH Domains of KSRP | | Descriptor: | Far upstream element-binding protein 2 | | Authors: | Diaz-Moreno, I, Hollingworth, D, Garcia-Mayoral, M.F, Kelly, G, Cukier, C.D, Ramos, A. | | Deposit date: | 2007-09-28 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Second and Third KH Domains of KSRP
To be Published, 2007
|
|
2MJH
 
 | |
2M32
 
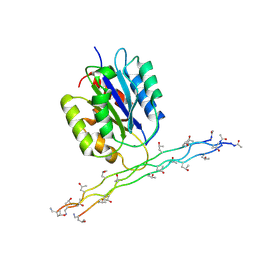 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
2K23
 
 | |
2M98
 
 | |
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
2MHB
 
 | |
2JXY
 
 | | Solution structure of the hemopexin-like domain of MMP12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M. | | Deposit date: | 2007-12-01 | | Release date: | 2008-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12
J.Am.Chem.Soc., 130, 2008
|
|
