8I1Y
 
 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8EI4
 
 | | Crystal structure of the WWP1 HECT domain in complex with H302, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, H302, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EIA
 
 | | Crystal structure of beta-catenin and the MDM2 p53-binding domain in complex with H333, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, E3 ubiquitin-protein ligase Mdm2, H333, ... | | Authors: | Li, K, Travaline, T.L, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8I2A
 
 | |
8ETY
 
 | |
8EL6
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 with an altered helix hA/hY conformation | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
5I52
 
 | |
8I5I
 
 | |
8I1W
 
 | |
5I4A
 
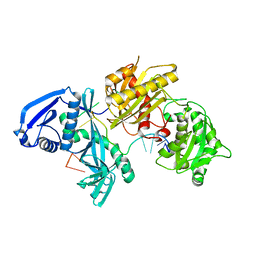 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | Descriptor: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | Authors: | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
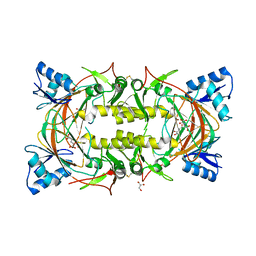
8EVN
 
 | |
8INH
 
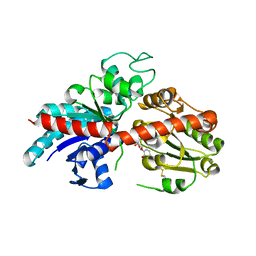 | | ZjOGT3, flavonoid 7,4'-di-O-glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, Wang, H.D, Li, F.D, Ye, M. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4'-di- O -glycosyltransferase from Ziziphus jujuba var. spinosa.
Chem Sci, 14, 2023
|
|
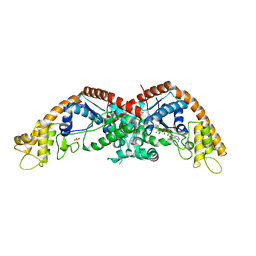
8I2C
 
 | |
8I5H
 
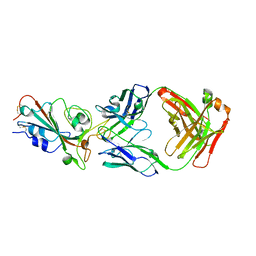 | |
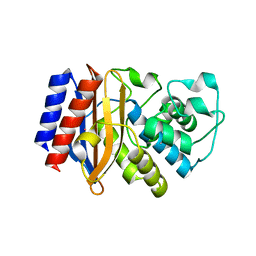
5I63
 
 | |
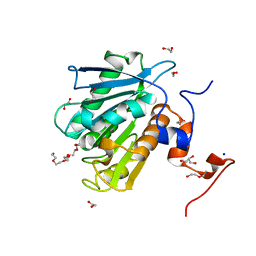
8ETZ
 
 | |
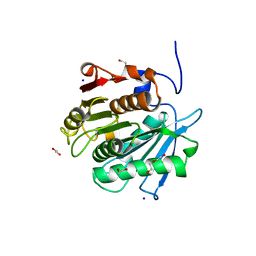
8EU0
 
 | |
8I1Z
 
 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I8Y
 
 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5I6X
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with paroxetine at the central site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
8I2L
 
 | |
8ETX
 
 | |
8ILZ
 
 | | Crystal structure of the RRM domain of human SETD1B | | Descriptor: | Histone-lysine N-methyltransferase SETD1B | | Authors: | Bao, S, Xu, C. | | Deposit date: | 2023-03-05 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular insight into the SETD1A/B N-terminal region and its interaction with WDR82.
Biochem.Biophys.Res.Commun., 658, 2023
|
|
8ILY
 
 | | Crystal structure of the RRM domain of human SETD1A | | Descriptor: | SET domain containing 1A, histone lysine methyltransferase | | Authors: | Bao, S, Xu, C. | | Deposit date: | 2023-03-05 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the SETD1A/B N-terminal region and its interaction with WDR82.
Biochem.Biophys.Res.Commun., 658, 2023
|
|
5I8Z
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT12 | | Descriptor: | 5-HEXYL-2-(4-NITROPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
