7M8M
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 3C-like proteinase, 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Z
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3-hydroxy-3-methylbutoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8N
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7AVI
 
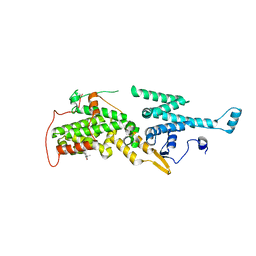 | | Crystal structure of SOS1 in complex with compound 2 | | Descriptor: | 3-propan-2-yl-~{N}-[(1~{R})-1-(3-sulfamoylphenyl)ethyl]-[1,2]oxazolo[5,4-b]pyridine-5-carboxamide, Son of sevenless homolog 1 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
3U57
 
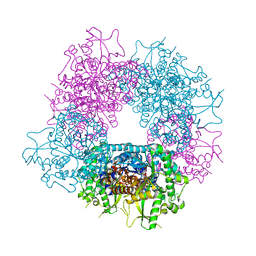 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
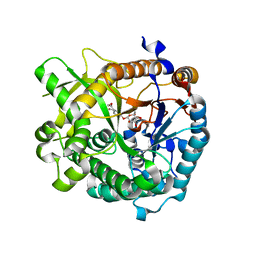 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
5U3W
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 7 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(cyclopropyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8F34
 
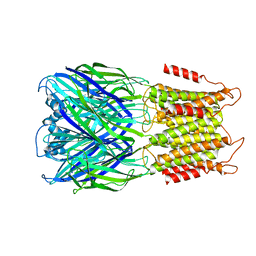 | | ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F33
 
 | | ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
3T6Y
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-{[(R)-(1-methyl-1H-imidazol-2-yl)(phenyl)methyl]amino}uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
8F32
 
 | | ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
8ESS
 
 | | Myoglobin variant Mb-cIII complex | | Descriptor: | Myoglobin, SULFATE ION, [2,18-bis(2-carboxyethyl)-7,12-diethenyl-3,8,13,17-tetramethyl-21-(2-oxo-3-phenylpropyl)porphyrin-21-iumato(2-)-kappa~3~N~22~,N~23~,N~24~]iron(2+) | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
1ONY
 
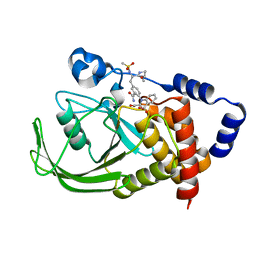 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
3U46
 
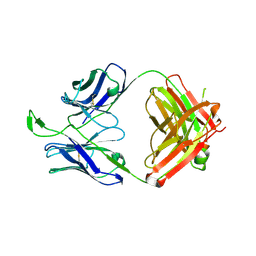 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
1ZZ3
 
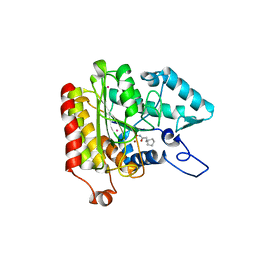 | | Crystal structure of a HDAC-like protein with CypX bound | | Descriptor: | 3-CYCLOPENTYL-N-HYDROXYPROPANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
8FE1
 
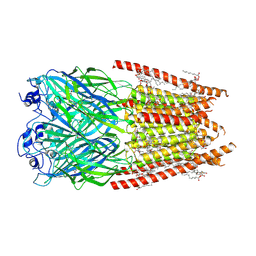 | | Alpha1/BetaB Heteromeric Glycine Receptor in 1 mM Glycine 20 uM Ivermectin State | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbs, E, Chakrapani, S. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor.
Nat Commun, 14, 2023
|
|
6DI4
 
 | |
1ONZ
 
 | | Oxalyl-aryl-Amino Benzoic acid Inhibitors of PTP1B, compound 8b | | Descriptor: | 2-[(7-HYDROXY-NAPHTHALEN-1-YL)-OXALYL-AMINO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
7CGL
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGI
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGK
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGF
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGG
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
