2KMC
 
 | | Solution Structure of the N-terminal domain of kindlin-1 | | Descriptor: | Fermitin family homolog 1 | | Authors: | Goult, B.T, Bate, N, Roberts, G.C, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the N-Terminus of Kindlin-1: A Domain Important for alphaIIbbeta3 Integrin Activation
J.Mol.Biol., 394, 2009
|
|
2KMS
 
 | | Combined high- and low-resolution techniques reveal compact structure in central portion of factor H despite long inter-modular linkers | | Descriptor: | Complement factor H | | Authors: | Schmidt, C.Q, Herbert, A.P, Guariento, M, Mertens, H.D.T, Soares, D.C, Uhrin, D, Rowe, A.J, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2009-08-04 | | Release date: | 2009-11-03 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | The Central Portion of Factor H (Modules 10-15) Is Compact and Contains a Structurally Deviant CCP Module
J.Mol.Biol., 395, 2010
|
|
2K59
 
 | |
2KEY
 
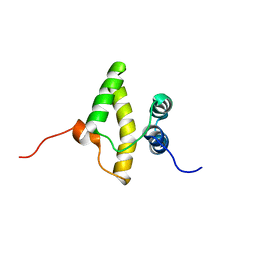 | | Solution NMR structure of a domain from a putative phage integrase protein BF2284 from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C | | Descriptor: | Putative phage integrase | | Authors: | Mills, J.L, Sathyamoorthy, B, Sukumaran, D.K, Lee, D, Ciccosanti, C, Jiang, M, Xiao, R, Acton, T.B, Swapna, G.V.T, Rost, B, Nair, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Domain from a Putative Phage Integrase Protein, BF2284, from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C
To be Published
|
|
2KH9
 
 | |
6N9E
 
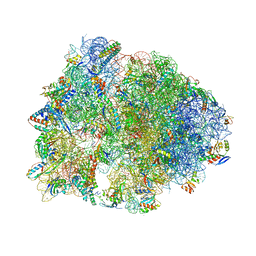 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6N9F
 
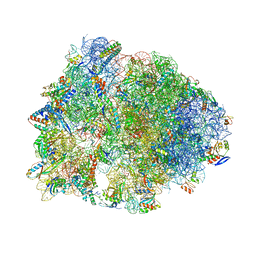 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
2KEN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of MM0293 from the archea Methanosarcina mazei
To be Published
|
|
2KBW
 
 | | Solution Structure of human Mcl-1 complexed with human Bid_BH3 peptide | | Descriptor: | BH3-interacting domain death agonist, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, Q, Moldoveanu, T, Sprules, T, Matta-Camacho, E, Mansur-Azzam, N, Gehring, K. | | Deposit date: | 2008-12-09 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Apoptotic regulation by MCL-1 through heterodimerization.
J.Biol.Chem., 285, 2010
|
|
2KPL
 
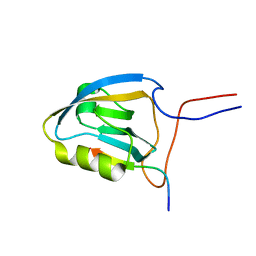 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2KTZ
 
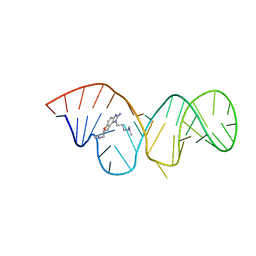 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KTU
 
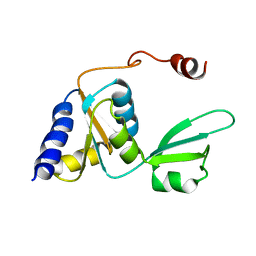 | |
2KCU
 
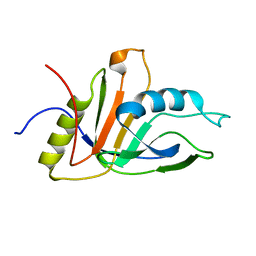 | | NMR solution structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107 | | Descriptor: | protein CtR107 | | Authors: | Mills, J.L, Zhang, Q, Sukumaran, D.K, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107
To be Published
|
|
7OKX
 
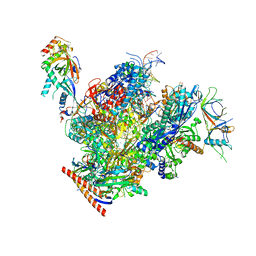 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5)-ELL2-EAF1 (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | Deposit date: | 2021-05-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
2KPN
 
 | | Solution NMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A | | Descriptor: | Bacillolysin, CALCIUM ION | | Authors: | Aramini, J.M, Wang, D, Ciccosanti, C.T, Janjua, H, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A
To be Published
|
|
2KWI
 
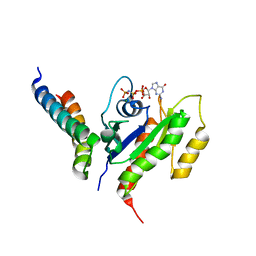 | | RalB-RLIP76 (RalBP1) complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RalA-binding protein 1, ... | | Authors: | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
2KMF
 
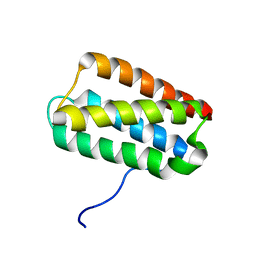 | | Solution Structure of Psb27 from cyanobacterial photosystem II | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Mabbitt, P.D, Rautureau, G.J.P, Day, C.L, Wilbanks, S.M, Eaton-Rye, J.J, Hinds, M.G. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Psb27 from cyanobacterial photosystem II
Biochemistry, 48, 2009
|
|
2KNY
 
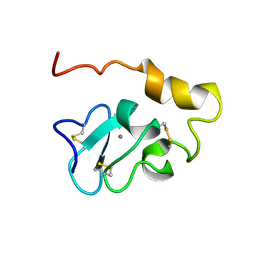 | |
2KMJ
 
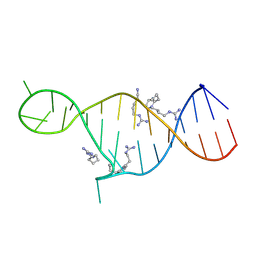 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
2KRE
 
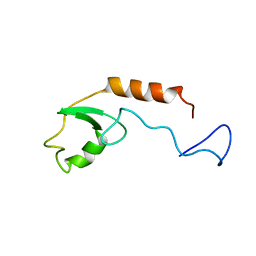 | |
2KSZ
 
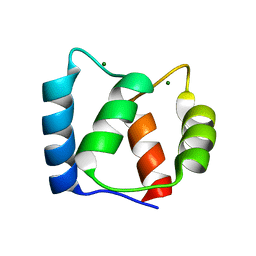 | |
5VJB
 
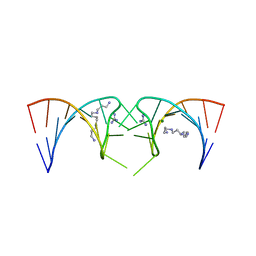 | |
2KXM
 
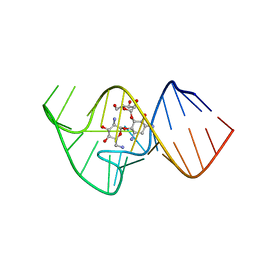 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7OOP
 
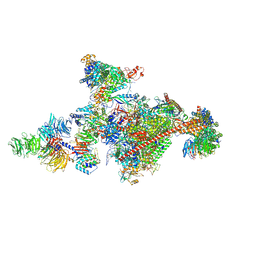 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OO3
 
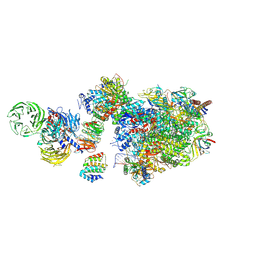 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
