7EEI
 
 | |
3OL9
 
 | |
3OL7
 
 | | Poliovirus polymerase elongation complex with CTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4ROE
 
 | | Human TFIIB-related factor 2 (Brf2) and TBP bound to RPPH1 promoter | | Descriptor: | MAGNESIUM ION, Non-template strand, TATA-box-binding protein, ... | | Authors: | Vannini, A, Gouge, J, Satia, K, Guthertz, N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2.
Cell(Cambridge,Mass.), 163, 2015
|
|
4ROC
 
 | | Human TFIIB-related factor 2 (Brf2) and TBP bound to U6#2 promoter | | Descriptor: | MAGNESIUM ION, Non-template strand, TATA-box-binding protein, ... | | Authors: | Vannini, A, Gouge, J, Satia, K, Guthertz, N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2.
Cell(Cambridge,Mass.), 163, 2015
|
|
3OL8
 
 | | Poliovirus polymerase elongation complex with CTP-Mn | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, MANGANESE (II) ION, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OL6
 
 | | Poliovirus polymerase elongation complex | | Descriptor: | ISOPROPYL ALCOHOL, Polymerase, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*UP*CP*CP*GP*GP*AP*AP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OLB
 
 | | Poliovirus polymerase elongation complex with 2',3'-dideoxy-ctp | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ISOPROPYL ALCOHOL, Polymerase, ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1NHV
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NHU
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
2WCX
 
 | |
5IHE
 
 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, ... | | Authors: | Sauguet, L, Raia, P, De Larue, M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography.
Nat Commun, 7, 2016
|
|
5IJL
 
 | |
7Z8Q
 
 | | Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Brodolin, K. | | Deposit date: | 2022-03-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Nat Commun, 14, 2023
|
|
2AWZ
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
8H2N
 
 | |
4OIN
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIO
 
 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6YHZ
 
 | |
6YI2
 
 | |
4OIP
 
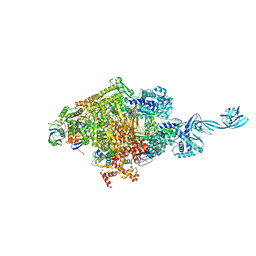 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5F3T
 
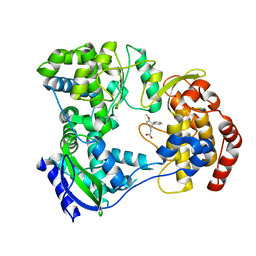 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to JF-31-MG46 | | Descriptor: | 2-(4-methoxy-3-phenyl-phenyl)ethanoic acid, RNA-DEPENDENT RNA POLYMERASE, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening.
J.Biol.Chem., 291, 2016
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
2AX0
 
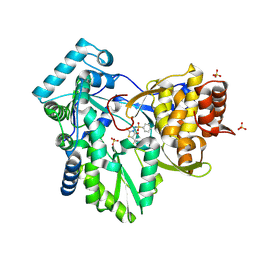 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
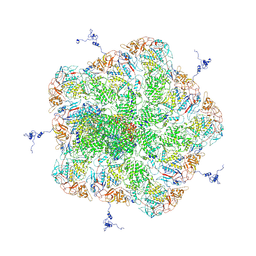
5ZVS
 
 | |
