7SDR
 
 | | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({methyl[(1R)-1-(naphthalen-1-yl)ethyl]amino}methyl)phenol, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor
To be Published
|
|
7NBN
 
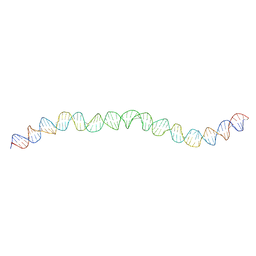 | | Allostery through DNA drives phenotype switching | | Descriptor: | AddAB promoter | | Authors: | Rosenblum, G, Elad, N, Rozenberg, H, Wiggers, F, Jungwirth, J, Hofmann, H. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allostery through DNA drives phenotype switching.
Nat Commun, 12, 2021
|
|
1M6X
 
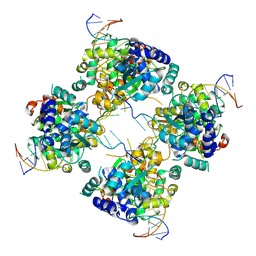 | | Flpe-Holliday Junction Complex | | Descriptor: | Flp recombinase, Symmetrized FRT site | | Authors: | Conway, A.B, Chen, Y, Rice, P.A. | | Deposit date: | 2002-07-17 | | Release date: | 2003-02-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Plasticity of the Flp-Holliday Junction Complex
J.Mol.Biol., 326, 2003
|
|
1DRG
 
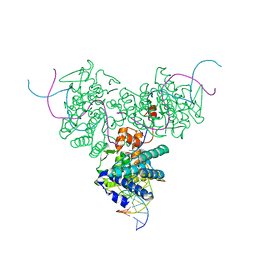 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | Descriptor: | 5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3', CRE RECOMBINASE | | Authors: | Woods, K.C, Baldwin, E.P. | | Deposit date: | 2000-01-06 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction
J.Mol.Biol., 313, 2001
|
|
4ZTT
 
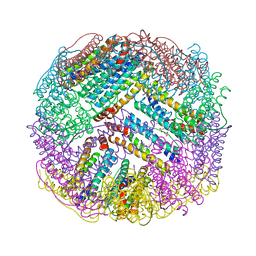 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
5A62
 
 | | Hydrolytic potential of the ammonia-oxidizing Thaumarchaeon Nitrososphaera gargenis - crystal structure and activity profiles of carboxylesterases linked to their metabolic function | | Descriptor: | ACETATE ION, PUTATIVE ALPHA/BETA HYDROLASE FOLD PROTEIN | | Authors: | Chow, J, Kaljunen, H, Nittinger, E, Spieck, E, Rarey, M, Mueller-Dieckmann, J, Streit, W.R. | | Deposit date: | 2015-06-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrolytic Potential of the Ammonia-Oxidizing Thaumarchaeon Nitrososphaera Gargenis - Crystal Structure and Activity Profiles of Carboxylesterases Linked to Their Metabolic Function
To be Published
|
|
5A3K
 
 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
1EA6
 
 | |
2LU0
 
 | | NMR solution structure of the kappa-zeta region of S.cerevisiae group II intron ai5(gamma) | | Descriptor: | RNA (49-MER) | | Authors: | Donghi, D, Pechlaner, M, Finazzo, C, Knobloch, B, Sigel, R.K.O. | | Deposit date: | 2012-06-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural stabilization of the kappa three-way junction by Mg(II) represents the first step in the folding of a group II intron.
Nucleic Acids Res., 41, 2013
|
|
7KP4
 
 | |
1EG6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D(CG(5-BRU)ACG) COMPLEXES TO A PHENAZINE | | Descriptor: | 5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3', 9-BROMO-PHENAZINE-1-CARBOXYLIC ACID (2-DIMETHYLAMINO-ETHYL)-AMIDE, BROMIDE ION, ... | | Authors: | Cardin, C.J, Denny, W.A, Hobbs, J.R, Thorpe, J.H. | | Deposit date: | 2000-02-14 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions.
Biochemistry, 39, 2000
|
|
6DUY
 
 | |
6LT4
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae: ATPrS-bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit, MAGNESIUM ION, ... | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
2R9A
 
 | | Crystal structure of human XLF | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Mol.Cell, 28
|
|
6LSY
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae - ATP bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.33 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
6M5Q
 
 | | A class C beta-lactamase mutant - Y150F | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
3CRX
 
 | |
6MHY
 
 | |
6MYE
 
 | | Crystal structure of human Scribble PDZ1 domain in complex with internal PDZ binding motif of Src homology 3 domain-containing guanine nucleotide exchange factor (SGEF) | | Descriptor: | FORMIC ACID, Protein scribble homolog, Rho guanine nucleotide exchange factor 26, ... | | Authors: | Sun, Y.J, Hou, T, Gakhar, L, Fuentes, E.J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | SGEF forms a complex with Scribble and Dlg1 and regulates epithelial junctions and contractility.
J.Cell Biol., 218, 2019
|
|
6MYF
 
 | |
7RUD
 
 | | DAHP synthase complex with trifluoropyruvate oxime | | Descriptor: | (2Z)-3,3,3-trifluoro-2-(hydroxyimino)propanoic acid, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive | | Authors: | Heimhalt, M, Mukherjee, P, Grainger, R, Szabla, R, Brown, C, Turner, R, Junop, M.S, Berti, P.J. | | Deposit date: | 2021-08-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Inhibitor-in-Pieces Approach to DAHP Synthase Inhibition: Potent Enzyme and Bacterial Growth Inhibition.
Acs Infect Dis., 7, 2021
|
|
7RUE
 
 | | DAHP synthase complexed with trifluoropyruvate semicarbazone | | Descriptor: | (2E)-2-(2-carbamoylhydrazinylidene)-3,3,3-trifluoropropanoic acid, MANGANESE (II) ION, Phospho-2-dehydro-3-deoxyheptonate aldolase, ... | | Authors: | Heimhalt, M, Mukherjee, P, Grainger, R, Szabla, R, Brown, C, Turner, R, Junop, M.S, Berti, P.J. | | Deposit date: | 2021-08-16 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Inhibitor-in-Pieces Approach to DAHP Synthase Inhibition: Potent Enzyme and Bacterial Growth Inhibition.
Acs Infect Dis., 7, 2021
|
|
2G88
 
 | | MSRECA-dATP COMPLEX | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Krishna, R, Manjunath, G.P, Kumar, P, Surolia, A, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery.
Nucleic Acids Res., 34, 2006
|
|
1F44
 
 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | Descriptor: | CRE RECOMBINASE, DNA (5'- D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3') | | Authors: | Baldwin, E.P, Woods, K.C. | | Deposit date: | 2000-06-07 | | Release date: | 2001-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction.
J.Mol.Biol., 313, 2001
|
|
5JK9
 
 | | Crystal structure of human IZUMO1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Izumo sperm-egg fusion protein 1 | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization
Nature, 534, 2016
|
|
