1FF7
 
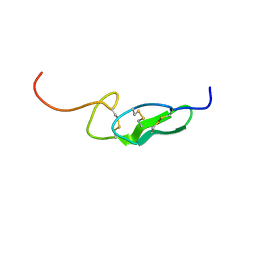 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII (FUCOSYLATED AT SER-60), NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (Blood Coagulation Factor VII), alpha-L-fucopyranose | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1B97
 
 | |
1B96
 
 | |
1F7M
 
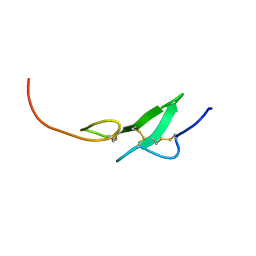 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (Blood Coagulation Factor VII) | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
1F7E
 
 | | THE FIRST EGF-LIKE DOMAIN FROM HUMAN BLOOD COAGULATION FVII, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (Blood Coagulation Factor VII) | | Authors: | Kao, Y.-H, Lee, G.F, Wang, Y, Starovasnik, M.A, Kelley, R.F, Spellman, M.W, Lerner, L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-06-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The effect of O-fucosylation on the first EGF-like domain from human blood coagulation factor VII.
Biochemistry, 38, 1999
|
|
4SKN
 
 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | Authors: | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-02-20 | | Release date: | 1999-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
1B98
 
 | | NEUROTROPHIN 4 (HOMODIMER) | | Descriptor: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-22 | | Release date: | 1999-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2CBR
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN I IN COMPLEX WITH A RETINOBENZOIC ACID (AM80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, PROTEIN (CRABP-I) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1FLC
 
 | | X-RAY STRUCTURE OF THE HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN OF INFLUENZA C VIRUS | | Descriptor: | HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rosenthal, P.B, Zhang, X, Formanowski, F, Fitz, W, Wong, C.H, Meier-Ewert, H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-02-22 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.
Nature, 396, 1998
|
|
1YLV
 
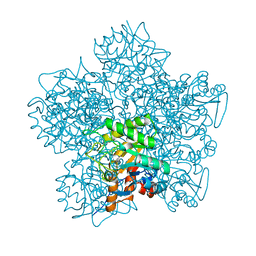 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID | | Descriptor: | LAEVULINIC ACID, PROTEIN (5-AMINOLAEVULINIC ACID DEHYDRATASE), ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Roper, J, Coker, A, Warren, M.J, Shoolingin-Jordan, P.M, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-02-22 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Schiff base complex of yeast 5-aminolaevulinic acid dehydratase with laevulinic acid.
Protein Sci., 8, 1999
|
|
1B99
 
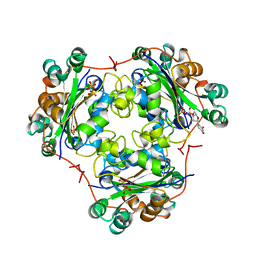 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1999-02-22 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
3CBS
 
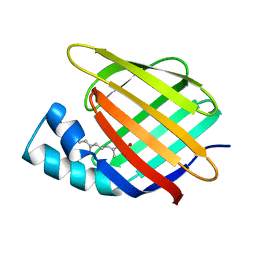 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-12 7310) | | Descriptor: | (2E,4E,6E,8E)-9-(4-hydroxy-2,3,6-trimethylphenyl)-3,7-dimethylnona-2,4,6,8-tetraenoic acid, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SBB
 
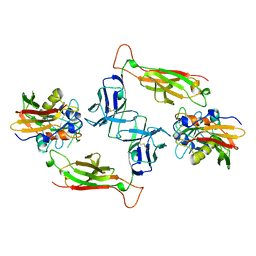 | | T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SUPERANTIGEN SEB | | Descriptor: | PROTEIN (14.3.D T CELL ANTIGEN RECEPTOR), PROTEIN (STAPHYLOCOCCAL ENTEROTOXIN B) | | Authors: | Li, H, Mariuzza, R.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B.
Immunity, 9, 1998
|
|
2UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE), ... | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
1NGL
 
 | | HUMAN NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (HNGAL), REGULARISED AVERAGE NMR STRUCTURE | | Descriptor: | PROTEIN (NGAL) | | Authors: | Coles, M, Diercks, T, Muehlenweg, B, Bartsch, S, Zoelzer, V, Tschesche, H, Kessler, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human neutrophil gelatinase-associated lipocalin.
J.Mol.Biol., 289, 1999
|
|
1JEN
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE | | Descriptor: | PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (ALPHA CHAIN)), PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (BETA CHAIN)) | | Authors: | Ekstrom, J.L, Mathews, I.I, Stanley, B.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 1999-02-23 | | Release date: | 1999-06-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold.
Structure Fold.Des., 7, 1999
|
|
1B93
 
 | | METHYLGLYOXAL SYNTHASE FROM ESCHERICHIA COLI | | Descriptor: | FORMIC ACID, PHOSPHATE ION, PROTEIN (METHYLGLYOXAL SYNTHASE) | | Authors: | Saadat, D, Harrison, D.H.T. | | Deposit date: | 1999-02-23 | | Release date: | 1999-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of methylglyoxal synthase from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1CA4
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 (TRAF2) | | Descriptor: | PROTEIN (TNF RECEPTOR ASSOCIATED FACTOR 2) | | Authors: | Park, Y.C, Burkitt, V, Villa, A.R, Tong, L, Wu, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for self-association and receptor recognition of human TRAF2.
Nature, 398, 1999
|
|
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1CAQ
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | Descriptor: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-23 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
1SHV
 
 | | STRUCTURE OF SHV-1 BETA-LACTAMASE | | Descriptor: | CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, PROTEIN (BETA-LACTAMASE SHV-1) | | Authors: | Kuzin, A.P, Nukaga, M, Nukaga, Y, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the SHV-1 beta-lactamase.
Biochemistry, 38, 1999
|
|
1TGO
 
 | | THERMOSTABLE B TYPE DNA POLYMERASE FROM THERMOCOCCUS GORGONARIUS | | Descriptor: | PROTEIN (THERMOSTABLE B DNA POLYMERASE) | | Authors: | Hopfner, K.-P, Eichinger, A, Engh, R.A, Laue, F, Ankenbauer, W, Huber, R, Angerer, B. | | Deposit date: | 1999-02-23 | | Release date: | 1999-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5LVE
 
 | |
