2YSA
 
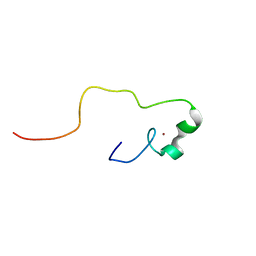 | | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1) | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Ohnishi, S, Sato, M, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1)
To be Published
|
|
6MWR
 
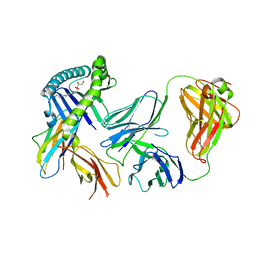 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
1N6U
 
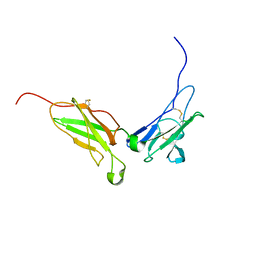 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
3IPT
 
 | |
7U2L
 
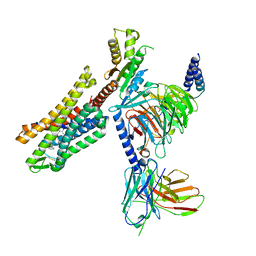 | | C5guano-uOR-Gi-scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Qu, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
2PEV
 
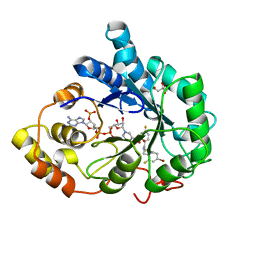 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution exceeds concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-03 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
4Z7G
 
 | |
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
1WMU
 
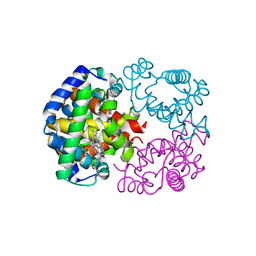 | | Crystal Structure of Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea, at 1.65 A resolution | | Descriptor: | Hemoglobin A and D beta chain, Hemoglobin D alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Satoh, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea, at 1.65 A resolution
To be Published
|
|
2XR6
 
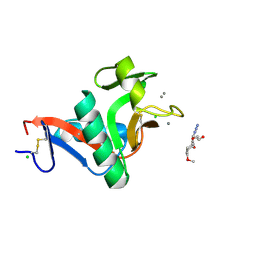 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
3MXT
 
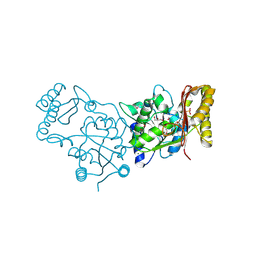 | | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni
To be Published
|
|
5MZU
 
 | |
2PFH
 
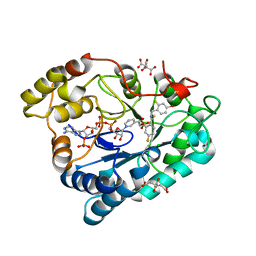 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is less than concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
4NXH
 
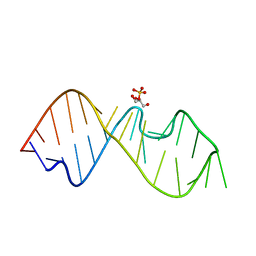 | |
3RBV
 
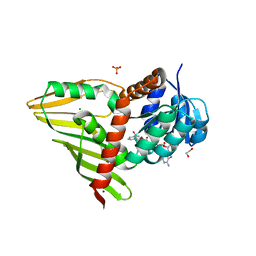 | | Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata incomplex with NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combined Structural and Functional Investigation of a C-3''-Ketoreductase Involved in the Biosynthesis of dTDP-l-Digitoxose.
Biochemistry, 50, 2011
|
|
2HCA
 
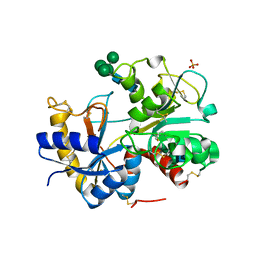 | | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Ethayathulla, A.S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution
To be Published
|
|
3B98
 
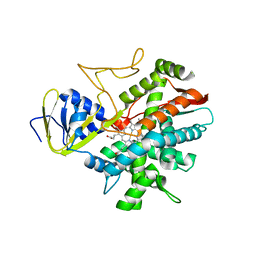 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
4PWL
 
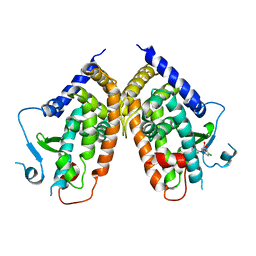 | | Crystal structure of the complex between PPARgamma-LBD and the S enantiomer of Mbx-102 (Metaglidasen) | | Descriptor: | (2S)-(4-chlorophenyl)[3-(trifluoromethyl)phenoxy]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A, Piemontese, L. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | On the metabolically active form of metaglidasen: improved synthesis and investigation of its peculiar activity on peroxisome proliferator-activated receptors and skeletal muscles.
Chemmedchem, 10, 2015
|
|
4I7K
 
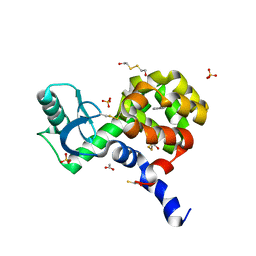 | | T4 Lysozyme L99A/M102H with toluene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
3RC2
 
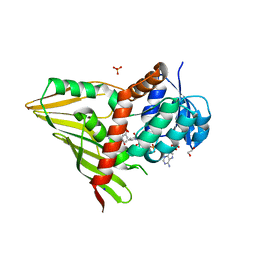 | | Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP; open conformation | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Functional Investigation of a C-3''-Ketoreductase Involved in the Biosynthesis of dTDP-l-Digitoxose.
Biochemistry, 50, 2011
|
|
3RCB
 
 | |
4I7Q
 
 | |
4I7L
 
 | | T4 Lysozyme L99A/M102H with phenol bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
3MUE
 
 | | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium | | Descriptor: | ACETIC ACID, ETHANOL, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium
To be Published
|
|
2Y61
 
 | | Crystal structure of Leishmanial E65Q-TIM complexed with S-Glycidol phosphate | | Descriptor: | GLYCEROL, SN-GLYCEROL-1-PHOSPHATE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Venkatesan, R, Alahuhta, M, Pihko, P.M, Wierenga, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High resolution crystal structures of triosephosphate isomerase complexed with its suicide inhibitors: the conformational flexibility of the catalytic glutamate in its closed, liganded active site.
Protein Sci., 20, 2011
|
|
