2MBS
 
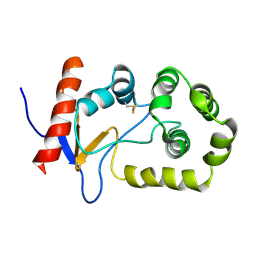 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
2NDO
 
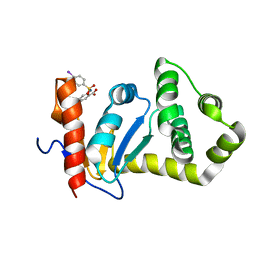 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
7L76
 
 | |
7LUI
 
 | |
7LUH
 
 | |
7LUJ
 
 | |
7LSM
 
 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
8D12
 
 | |
8CZM
 
 | |
8CXD
 
 | |
8CZN
 
 | |
8CXE
 
 | |
8D10
 
 | |
8D11
 
 | |
7L7C
 
 | |
7LHP
 
 | |
4ZL7
 
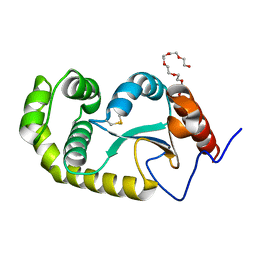 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal I | | Descriptor: | HEXAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL8
 
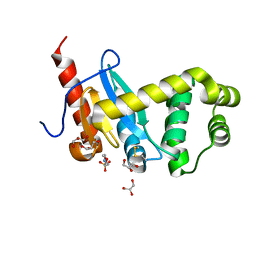 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | McMahoh, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZL9
 
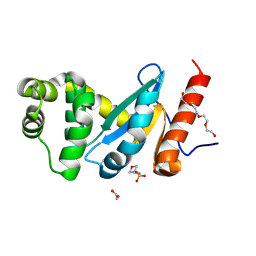 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I: Crystal III | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-05-01 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sent packing: protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4ZIJ
 
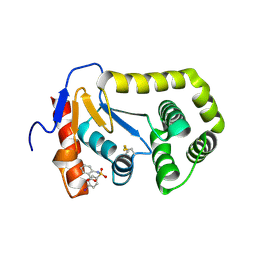 | | Crystal structure of E.Coli DsbA in complex with 2-(4-iodophenylsulfonamido) benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Vazirani, M, Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
8DG0
 
 | |
8DG1
 
 | |
8DG2
 
 | | Crystal Structure of EcDsbA in a complex with DMSO | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Whitehouse, R.L, Ilyichova, O.V, Taylor, A.J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment screening libraries for the identification of protein hot spots and their minimal binding pharmacophores.
Rsc Med Chem, 14, 2023
|
|
8DN0
 
 | |
1A2J
 
 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
