4WIA
 
 | |
6A7F
 
 | |
5DHM
 
 | |
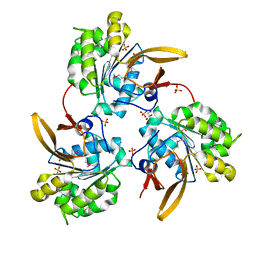
7LQ4
 
 | | Rr (RsiG)2-(c-di-GMP)2-WhiG complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), RsiG, WhiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
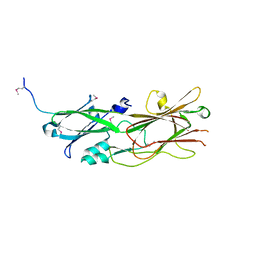
7LQ2
 
 | | Apo Rr RsiG- crystal form 1 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, RR RsiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LQ3
 
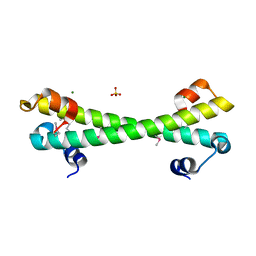 | |
1J8S
 
 | | PAPG ADHESIN RECEPTOR BINDING DOMAIN-UNBOUND FORM | | Descriptor: | PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G. | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
8CYE
 
 | |
8CWM
 
 | |
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
8CVI
 
 | |
1R8I
 
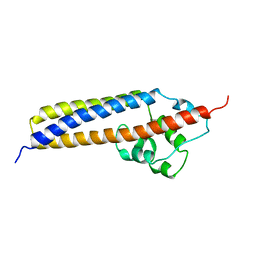 | | Crystal structure of TraC | | Descriptor: | TraC | | Authors: | Yeo, H.-J, Yuan, Q, Beck, M.R, Baron, C, Waksman, G. | | Deposit date: | 2003-10-24 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5VLY
 
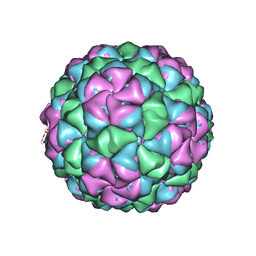 | | Asymmetric unit for the coat proteins of phage Qbeta | | Descriptor: | Capsid protein | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2KID
 
 | | Solution Structure of the S. Aureus Sortase A-substrate Complex | | Descriptor: | (PHQ)LPA(B27) peptide, CALCIUM ION, Sortase | | Authors: | Suree, N, Liew, C.K, Villareal, V.A, Thieu, W, Fadeev, E.A, Clemens, J.J, Jung, M.E, Clubb, R.T. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized.
J.Biol.Chem., 284, 2009
|
|
1J8R
 
 | | BINARY COMPLEX OF THE PAPG RECEPTOR-BINDING DOMAIN BOUND TO GBO4 RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
7BHD
 
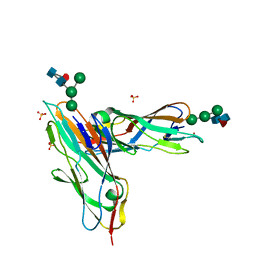 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
2YJL
 
 | | Structural characterization of a secretin pilot protein from the type III secretion system (T3SS) of Pseudomonas aeruginosa | | Descriptor: | EXOENZYME S SYNTHESIS PROTEIN B, NICKEL (II) ION, SULFATE ION | | Authors: | Izore, T, Perdu, C, Job, V, Atree, I, Faudry, E, Dessen, A. | | Deposit date: | 2011-05-20 | | Release date: | 2011-08-10 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Characterization and Membrane Localization of Exsb from the Type III Secretion System (T3Ss) of Pseudomonas Aeruginosa
J.Mol.Biol., 413, 2011
|
|
6YHW
 
 | | Co-crystals in the P212121 space group, of a beta-cyclodextrin spacered by triazole heptyl from alpha-D-mannose, with FimH lectin at 2.00 A resolution. | | Descriptor: | 2H-1,2,3-TRIAZOL-4-YLMETHANOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), FimH, ... | | Authors: | de Ruyck, J, Bouckaert, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The Antiadhesive Strategy in Crohn's Disease: Orally Active Mannosides to Decolonize Pathogenic Escherichia coli from the Gut.
Chembiochem, 17, 2016
|
|
6XL0
 
 | | Caulobacter crescentus FljK filament | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
6XKY
 
 | | Caulobacter crescentus FljK filament, straightened | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
4RTS
 
 | |
4RTR
 
 | |
4RTO
 
 | |
8B3O
 
 | |
5NFI
 
 | | The fimbrial anchor protein Mfa2 from Porphyromonas gingivalis | | Descriptor: | IODIDE ION, Minor fimbrium anchoring subunit Mfa2 | | Authors: | Hall, M, Hasegawa, Y, Persson, K, Yoshimura, F. | | Deposit date: | 2017-03-14 | | Release date: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis.
Sci Rep, 8, 2018
|
|
