4KL6
 
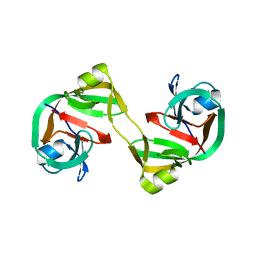 | | Crystal structure of dimeric form of NpuDnaE intein | | Descriptor: | DNA-directed DNA polymerase,Nucleic acid binding, OB-fold, tRNA/helicase-type | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
8J7S
 
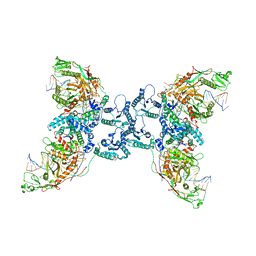 | | Structure of the SPARTA complex | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*CP*AP*AP*TP*AP*GP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*A)-3'), ... | | Authors: | Guo, M, Zhu, Y, Lin, Z, Huang, Z. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the ssDNA-activated SPARTA complex.
Cell Res., 33, 2023
|
|
6WHM
 
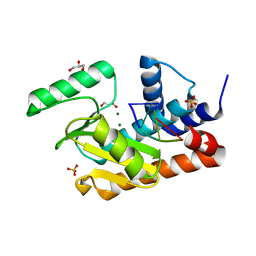 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomer TAGC (cleaved TTAGCATT, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*C)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
8DVI
 
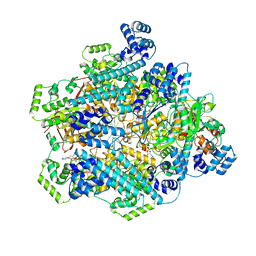 | | T4 bacteriophage primosome with single strand DNA, State 2 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (70-MER), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DVF
 
 | | T4 Bacteriophage primosome with single strand DNA, state 1 | | Descriptor: | DNA (5'-D(P*GP*GP*CP*TP*G)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DW6
 
 | | T4 bacteriophage primosome with single-strand DNA, State 3 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (70-MER), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DTP
 
 | | Close state of T4 bacteriophage gp41 hexamer bound with single strand DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DnaB-like replicative helicase, MAGNESIUM ION, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DUE
 
 | |
6WS3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomers TG and AGCA (from cleaved GTGAGCAGTG) | | Descriptor: | DNA (5'-D(P*TP*G)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
5B7J
 
 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
5KL6
 
 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
7V9U
 
 | | Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom | | Descriptor: | DNA (105-MER), RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*G)-3'), RNA (81-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
5NR3
 
 | | Human DNMT3B PWWP domain in complex with ethambutol | | Descriptor: | DNA (cytosine-5)-methyltransferase 3B, Ethambutol, SULFATE ION | | Authors: | Rondelet, G, Wouters, J. | | Deposit date: | 2017-04-21 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting PWWP domain of DNA methyltransferase 3B for epigenetic cancer therapy: Identification and structural characterization of new potential protein-protein interaction inhibitors
To be published
|
|
4TR8
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
2LZN
 
 | |
7X7P
 
 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7YDW
 
 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
8TQV
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20403 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N-{[1-(methoxymethyl)cyclopropyl]methyl}-N-methylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TRY
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20348 | | Descriptor: | N-{(2S,3S)-4-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-3-hydroxy-1-phenylbutan-2-yl}-4-(2-methylbutan-2-yl)benzene-1-sulfonamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-10 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TQG
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20419 | | Descriptor: | N-benzyl-2-{4-[4-(4,5-dimethoxy-1H-indole-2-carbonyl)piperazine-1-carbonyl]piperidin-1-yl}-6-methylpyrimidine-4-carboxamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TR4
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20404 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N,N-dimethylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
1DAU
 
 | | Analog of dickerson-drew DNA dodecamer with 6'-alpha-methyl carbocyclic thymidines, NMR, minimized average structure | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(T32)P*(T32)P*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1EG2
 
 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
6M4J
 
 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
