1EMH
 
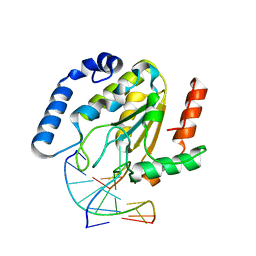 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO UNCLEAVED SUBSTRATE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(P2U)P*AP*TP*CP*TP*T)-3'), URACIL-DNA GLYCOSYLASE | | Authors: | Parikh, S.S, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DNY
 
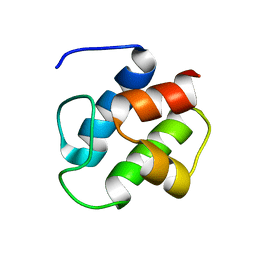 | | SOLUTION STRUCTURE OF PCP, A PROTOTYPE FOR THE PEPTIDYL CARRIER DOMAINS OF MODULAR PEPTIDE SYNTHETASES | | Descriptor: | NON-RIBOSOMAL PEPTIDE SYNTHETASE PEPTIDYL CARRIER PROTEIN | | Authors: | Weber, T, Baumgartner, R, Renner, C, Marahiel, M.A, Holak, T.A. | | Deposit date: | 1999-12-17 | | Release date: | 2000-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
Structure Fold.Des., 8, 2000
|
|
1EK6
 
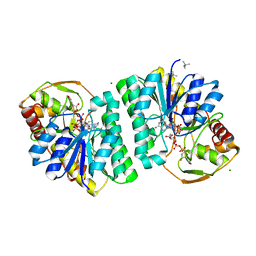 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH NADH AND UDP-GLUCOSE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, TETRAMETHYLAMMONIUM ION, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
1EWP
 
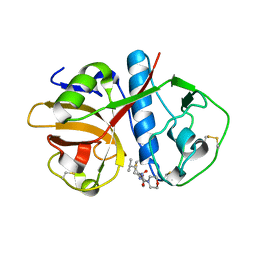 | | CRUZAIN BOUND TO MOR-LEU-HPQ | | Descriptor: | CRUZAIN, N-[(3S)-1-fluoro-2-oxo-5-phenylpentan-3-yl]-N~2~-(morpholin-4-ylcarbonyl)-L-leucinamide | | Authors: | Gillmor, S.A. | | Deposit date: | 2000-04-26 | | Release date: | 2000-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chapter 3: X-ray Structures of Complexes of Cruzain with Designed Covalent Inhibitors
Enzyme-ligand Interactions, Inhibition and Specificity, 1998
|
|
1MR8
 
 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EX3
 
 | |
1EHW
 
 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EK5
 
 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE IN COMPLEX WITH NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE 4-EPIMERASE | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
1CM8
 
 | | PHOSPHORYLATED MAP KINASE P38-GAMMA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PHOSPHORYLATED MAP KINASE P38-GAMMA | | Authors: | Bellon, S, Fitzgibbon, M.J, Fox, T, Hsiao, H.M, Wilson, K.P. | | Deposit date: | 1999-05-17 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation.
Structure Fold.Des., 7, 1999
|
|
1QC5
 
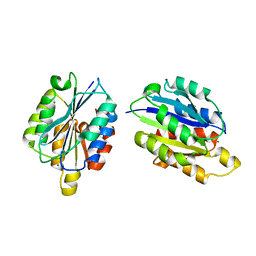 | | I Domain from Integrin Alpha1-Beta1 | | Descriptor: | MAGNESIUM ION, PROTEIN (ALPHA1 BETA1 INTEGRIN) | | Authors: | Deivanayagam, C.C, Narayana, S.V. | | Deposit date: | 1999-05-17 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM.
J.Biol.Chem., 274, 1999
|
|
1E1Y
 
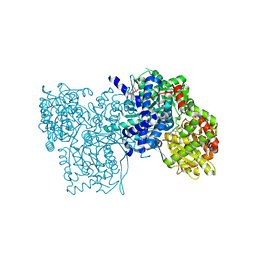 | | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Flavopiridol Inhibits Glycogen Phosphorylase by Binding at the Inhibitor Site
J.Biol.Chem., 275, 2000
|
|
1EYV
 
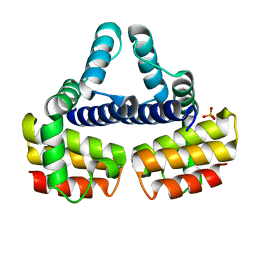 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1HD0
 
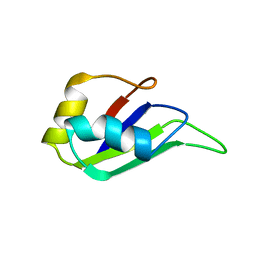 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
1HD1
 
 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
2MST
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | Descriptor: | PROTEIN (MUSASHI1) | | Authors: | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
1DKE
 
 | | NI BETA HEME HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN: ALPHA CHAIN, HEMOGLOBIN: BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bruno, S, Bettatti, S, Mozzarelli, A, Bolognesi, M, Deriu, D, Rosano, C, Tsuneshige, A, Yonetani, T, Henry, E.R. | | Deposit date: | 1999-12-07 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oxygen binding by alpha(Fe2+)2beta(Ni2+)2 hemoglobin crystals.
Protein Sci., 9, 2000
|
|
1DLQ
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1EHI
 
 | | D-ALANINE:D-LACTATE LIGASE (LMDDL2) OF VANCOMYCIN-RESISTANT LEUCONOSTOC MESENTEROIDES | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE:D-LACTATE LIGASE, ... | | Authors: | Kuzin, A.P, Sun, T, Jorczak-Baillass, J, Healy, V.L, Walsh, C.T, Knox, J.R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Enzymes of vancomycin resistance: the structure of D-alanine-D-lactate ligase of naturally resistant Leuconostoc mesenteroides.
Structure Fold.Des., 8, 2000
|
|
3LRI
 
 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
1DMH
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLM
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLT
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1QDE
 
 | |
