4WBY
 
 | | tRNA-processing enzyme (apo form I) | | Descriptor: | GLYCEROL, Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WC0
 
 | | tRNA-processing enzyme with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Poly A polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WBZ
 
 | | tRNA-processing enzyme (apo form 2) | | Descriptor: | Poly A polymerase | | Authors: | Yamashita, S, Tomtia, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
3NH2
 
 | |
3NH0
 
 | |
3NGZ
 
 | |
3NH1
 
 | |
3NGY
 
 | | Crystal structure of RNase T (E92G mutant) | | Descriptor: | COBALT (II) ION, Ribonuclease T, his tag sequence | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2010-06-14 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation
Nat.Chem.Biol., 7, 2011
|
|
8IUE
 
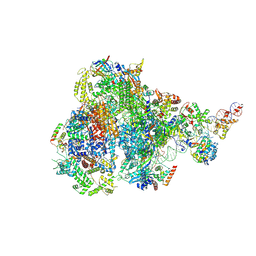 | | RNA polymerase III pre-initiation complex melting complex 1 | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8IUH
 
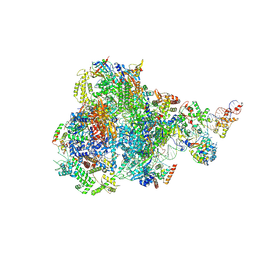 | | RNA polymerase III pre-initiation complex open complex 1 | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
5LQW
 
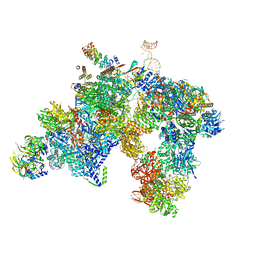 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
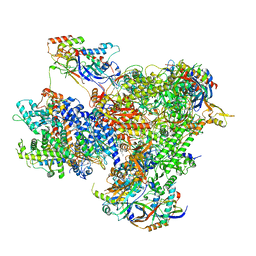 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
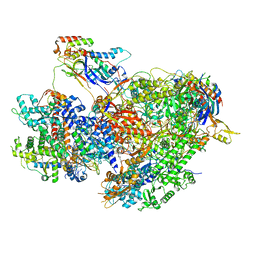 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
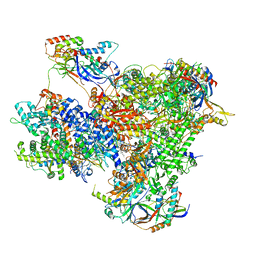 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
8EYQ
 
 | | 30S_delta_ksgA_h44_inactive_conformation | | Descriptor: | 16S_rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Ortega, J, Sun, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5A7Z
 
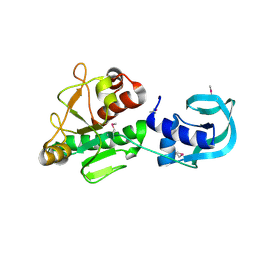 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.1 angstrom resolution. | | Descriptor: | TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | Authors: | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
5VVR
 
 | |
7VF5
 
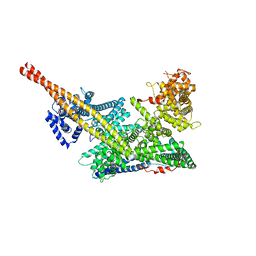 | | Human m6A-METTL associated complex (WTAP, VIRMA, and HAKAI) | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog | | Authors: | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
7VF2
 
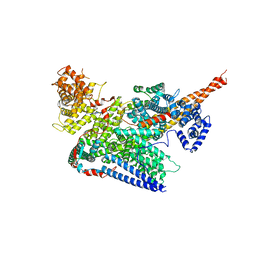 | | Human m6A-METTL associated complex (WTAP, VIRMA, ZC3H13, and HAKAI) | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog, Zinc finger CCCH domain-containing protein 13 | | Authors: | Su, S, Li, S, Deng, T, Gao, M, Yin, Y, Wu, B, Peng, C, Liu, J, Ma, J, Zhang, K. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human m6A writer complexes.
Cell Res., 32, 2022
|
|
8ODP
 
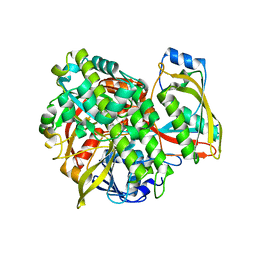 | | Structure of human RTCB with GMPCPP in complex with Archease | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Protein archease, ... | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
8ODO
 
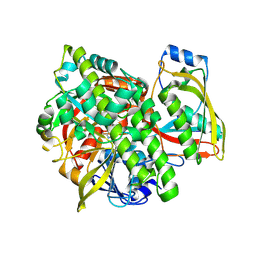 | | Structure of human guanylylated RTCB in complex with Archease | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
8BTT
 
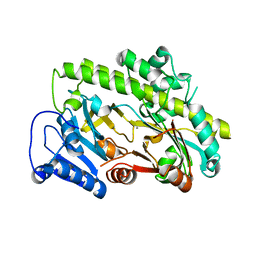 | | Structure of human RTCB | | Descriptor: | MANGANESE (II) ION, RNA-splicing ligase RtcB homolog | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
6L7V
 
 | |
6L7X
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #951 ligand | | Descriptor: | 1,3-dimethyl-7-propyl-purine-2,6-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
