7GGF
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6d04362c-2 (Mpro-x12423) | | Descriptor: | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-[4-(dimethylamino)phenyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-92e193ae-1 (Mpro-x12692) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGV
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-91acba05-6 (Mpro-x12710) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGX
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-971238d3-5 (Mpro-x12716) | | Descriptor: | (4R)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8DS9
 
 | | LRRC8A:C in MSPE3D1 nanodisc top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7GH6
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-916-2 (Mpro-x2910) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHB
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with GAB-REV-70cc3ca5-8 (Mpro-x3080) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N~2~-methyl-N-(4-methylpyridin-3-yl)-N~2~-(quinoline-8-sulfonyl)glycinamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8DR8
 
 | | LRRC8A:C conformation 2 (oblong) top mask | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7GHD
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-4e090d3a-57 (Mpro-x3298) | | Descriptor: | (2S)-N-{2-[(4-fluorobenzene-1-sulfonyl)amino]phenyl}-2-hydroxy-2-(pyridin-3-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with GAB-REV-70cc3ca5-13 (Mpro-x3108) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-N~2~-[(pyridin-3-yl)acetyl]glycinamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-2 (Mpro-x10334) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-8 (Mpro-x10423) | | Descriptor: | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8DRE
 
 | | LRRC8A:C conformation 2 (oblong) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7GCE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-93268d01-7 (Mpro-x10598) | | Descriptor: | 3-(2-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCV
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-2eddb1ff-7 (Mpro-x10789) | | Descriptor: | 2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8DRK
 
 | | LRRC8A:C conformation 1 (round) top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NT0
 
 | | Catalase 3 from N.Crassa in ferrous state, X-ray reduced (1.315 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
7GD2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-3 (Mpro-x10862) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-1-(3-bromophenyl)-2-methoxyethyl]-2-[(3S)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8DSA
 
 | | LRRC8A:C in MSP1E3D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DS3
 
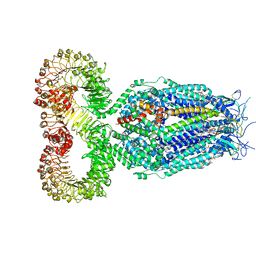 | | LRRC8A:C conformation 1 (round) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5IFS
 
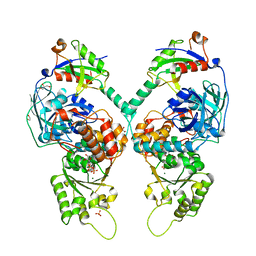 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
6S4H
 
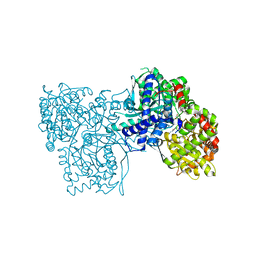 | | The crystal structure of glycogen phosphorylase in complex with 8 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6O1D
 
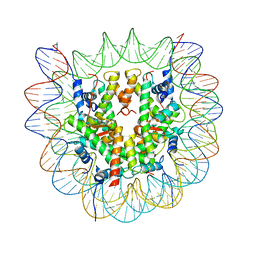 | |
