8HFH
 
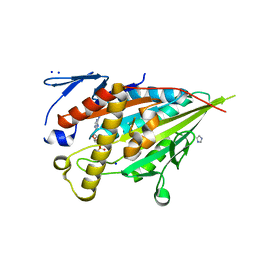 | | CENP-E motor domain in complex with AMPPNP and Mg2+ | | Descriptor: | Centromere-associated protein E, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Shibuya, A, Yokoyama, H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the motor domain of centromere-associated protein E in complex with a non-hydrolysable ATP analogue.
Febs Lett., 597, 2023
|
|
8F4S
 
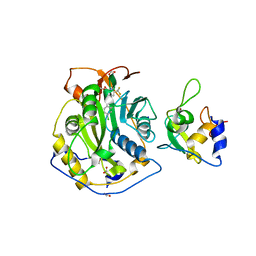 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
3RNX
 
 | |
8EU1
 
 | |
3QVI
 
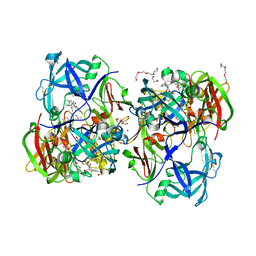 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
8ETY
 
 | |
1IKQ
 
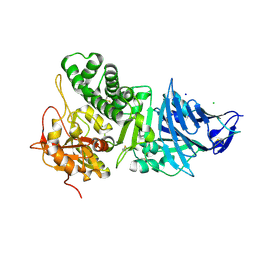 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
8ETK
 
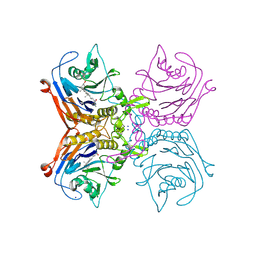 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-(2-{2-[(prop-2-yn-1-yl)oxy]ethoxy}ethoxy)hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Grundy, M.K, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
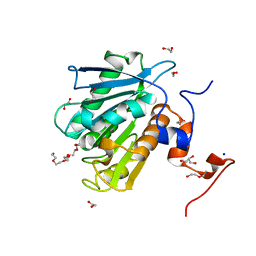
8ETZ
 
 | |
8EU0
 
 | |
3QSR
 
 | |
8ETX
 
 | |
8EWT
 
 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-{(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-[(prop-2-yn-1-yl)oxy]hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl}-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
3QVW
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus mutant K278A | | Descriptor: | GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
3QXW
 
 | | Free structure of an anti-methotrexate CDR1-4 Graft VHH Antibody | | Descriptor: | Anti-Methotrexate CDR1-4 Graft VHH, SODIUM ION, SULFATE ION | | Authors: | Fanning, S.W, Horn, J.R. | | Deposit date: | 2011-03-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop.
Protein Sci., 20, 2011
|
|
8HAP
 
 | | Crystal structure of thermostable acetaldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, SODIUM ION, ... | | Authors: | Mine, S, Nakabayashi, M, Ishikawa, K. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable acetaldehyde dehydrogenase from the hyperthermophilic archaeon Sulfolobus tokodaii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8EYP
 
 | | Joint X-ray/neutron structure of Salmonella typhimurium tryptophan synthase internal aldimine from microgravity-grown crystal | | Descriptor: | SODIUM ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Drago, V.N, Kovalevsky, A, Blakeley, M.P, Forsyth, V.T, Mueser, T.C. | | Deposit date: | 2022-10-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Neutron diffraction from a microgravity-grown crystal reveals the active site hydrogens of the internal aldimine form of tryptophan synthase.
Cell Rep Phys Sci, 5, 2024
|
|
8EZC
 
 | |
3Q4W
 
 | | The structure of archaeal inorganic pyrophosphatase in complex with substrate | | Descriptor: | BROMIDE ION, CALCIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Hughes, R.C, Meehan, E.J, Coates, L, Ng, J.D. | | Deposit date: | 2010-12-24 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Inorganic pyrophosphatase crystals from Thermococcus thioreducens for X-ray and neutron diffraction.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3PK4
 
 | | Urate oxidase under 3.2 MPa / 32 bars pressure of xenon | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
8H9O
 
 | |
3PKG
 
 | | Urate oxidase under 2 MPa / 20 bars pressure of xenon | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
8H9X
 
 | |
