6X1A
 
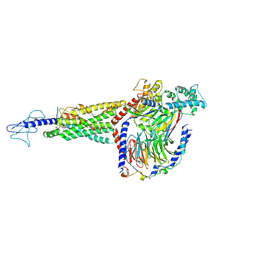 | | Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X0S
 
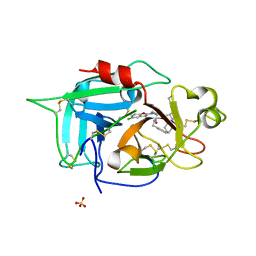 | |
6X0T
 
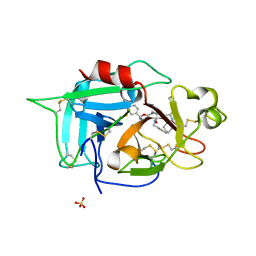 | | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h) | | Descriptor: | (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide, Coagulation factor XII, SULFATE ION | | Authors: | Orth, P. | | Deposit date: | 2020-05-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h)
To Be Published
|
|
6Z2C
 
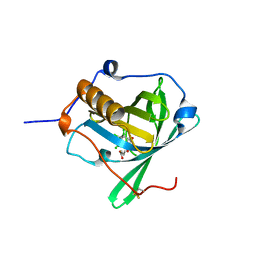 | | Engineered lipocalin C3A5 in complex with a transition state analog | | Descriptor: | 1,7,8,9,10,10-hexachloro-4-carboxypentyl-4-aza-tricyclo[5.2.1.0(2,6)]dec-8-ene-3,5-dione, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an engineered lipocalin that catalyzes a Diels-Alder reaction
To be published
|
|
6X0B
 
 | |
6Z29
 
 | | Structure of eIF4G1 (37-49) - PUB1 RRM3 chimera in solution | | Descriptor: | Eukaryotic initiation factor 4F subunit p150,Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Chaves-Arquero, B, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | eIF4G1 N-terminal intrinsically disordered domain is a multi-docking station for RNA, Pab1, Pub1, and self-assembly.
Front Mol Biosci, 9, 2022
|
|
7C4C
 
 | |
7C4B
 
 | | The crystal structure of Trypanosoma brucei RNase D : UMP complex | | Descriptor: | CCHC-type domain-containing protein, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C45
 
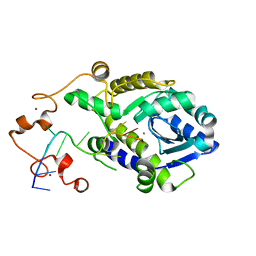 | |
7C47
 
 | | The crystal structure of Trypanosoma brucei RNase D : CMP complex | | Descriptor: | CCHC-type domain-containing protein, CYTIDINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
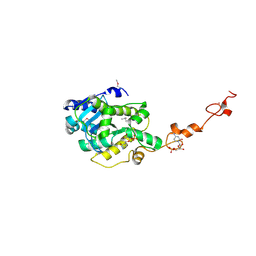
6Z1Q
 
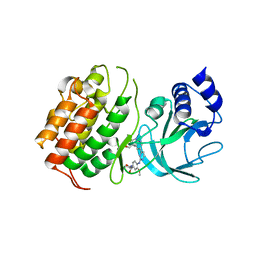 | | MAP3K14 (NIK) in complex with DesF-3R/4076 | | Descriptor: | DesF-3R/4076, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6WZM
 
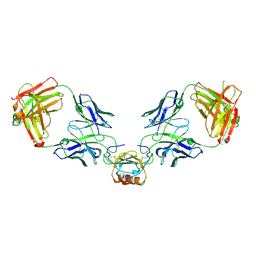 | | LY3041658 Fab bound to CXCL8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-8, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6WZJ
 
 | | LY3041658 Fab bound to CXCL2 | | Descriptor: | C-X-C motif chemokine 2, LY3041658 Fab Heavy Chain, LY3041658 Fab Light Chain | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6Z1U
 
 | | bovine ATP synthase F1c8-peripheral stalk domain, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z1R
 
 | | bovine ATP synthase F1-peripheral stalk domain, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZP
 
 | | The crystal structure of 4-vinylbenzoate-bound T252A mutant CYP199A4 | | Descriptor: | 4-ethenylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding the Mechanistic Requirements for Efficient and Stereoselective Alkene Epoxidation by a Cytochrome P450 Enzyme
Acs Catalysis, 11, 2021
|
|
7C40
 
 | | MgGDP bound KRAS G12V | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
6WZL
 
 | | LY3041658 Fab bound to CXCL7 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, C-X-C motif chemokine 7, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
6Z1T
 
 | | MAP3K14 (NIK) in complex with 4S/3694 | | Descriptor: | 4S/3694, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6WZK
 
 | | LY3041658 Fab bound to CXCL3 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C-X-C motif chemokine 3, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
7C43
 
 | |
7C42
 
 | |
7C41
 
 | | KRAS G12V and H-REV107 peptide complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, HRAS-like suppressor 3, ... | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
6Z1G
 
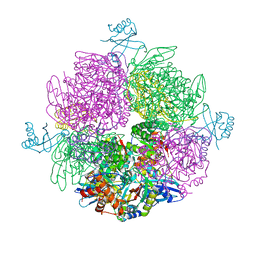 | | CryoEM structure of the interaction between Rubisco Activase small-subunit-like (SSUL) domain with Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, Ribulose bisphosphate carboxylase/oxygenase activase | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6Z19
 
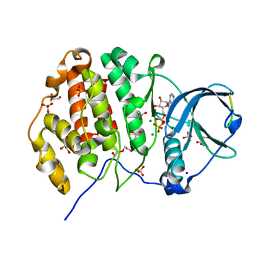 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
