1COW
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1CY7
 
 | |
1CY6
 
 | |
5DXB
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Stapled Peptide SRC2-P1 and Estradiol | | Descriptor: | CHLORIDE ION, ESTRADIOL, Estrogen receptor, ... | | Authors: | Fanning, S.W, Speltz, T.E, Mayne, C.G, Tajkhorshid, E, Greene, G.L, Moore, T.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-07-27 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Stapled Peptides with gamma-Methylated Hydrocarbon Chains for the Estrogen Receptor/Coactivator Interaction.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5H9E
 
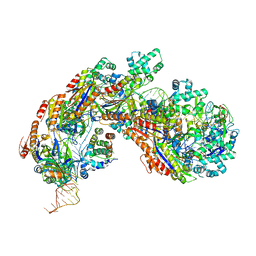 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
8EHC
 
 | |
6O18
 
 | |
6WUA
 
 | |
5KNB
 
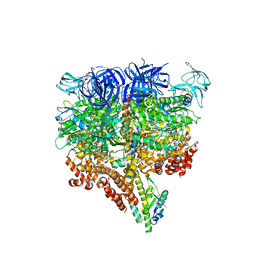 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
8BD7
 
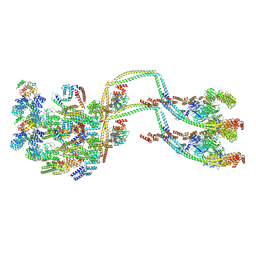 | | IFTB1 subcomplex of anterograde Intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Clusterin-associated protein 1, IFT54, IFT70, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EHS
 
 | |
6NZ8
 
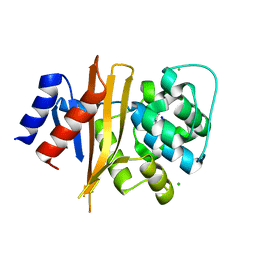 | | Structure of carbamylated apo OXA-231 carbapenemase | | Descriptor: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | Authors: | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
8F1B
 
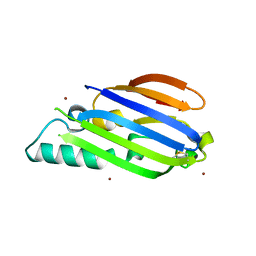 | |
8EZW
 
 | |
4V2E
 
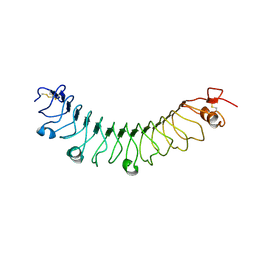 | | FLRT3 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 3 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development.
Neuron, 84, 2014
|
|
6O2E
 
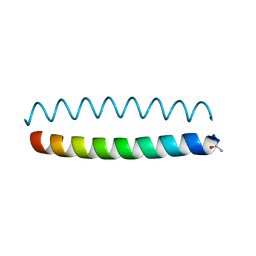 | |
6O2F
 
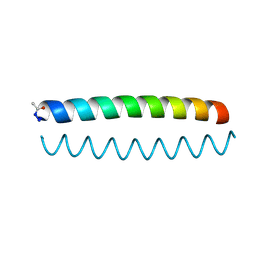 | |
7M8V
 
 | | Human CYP11B2 in complex with LCI699 | | Descriptor: | 4-[(4R,5R)-6,7-dihydro-5H-pyrrolo[1,2-c]imidazol-5-yl]-3-fluorobenzonitrile, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Aldosterone Synthase Structure With Cushing Disease Drug LCI699 Highlights Avenues for Selective CYP11B Drug Design.
Hypertension, 78, 2021
|
|
6WU9
 
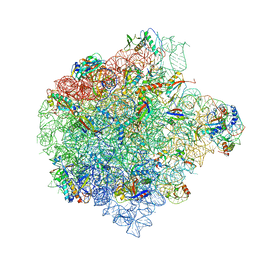 | |
5KND
 
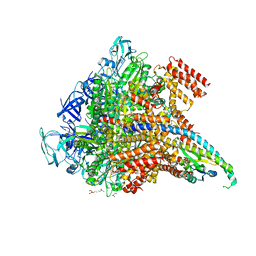 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
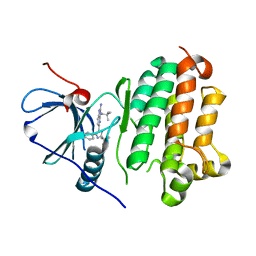
8G63
 
 | |
8G83
 
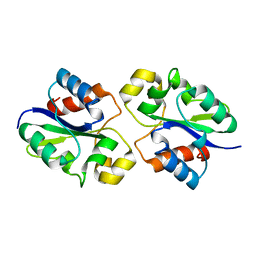 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
6OJW
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 holo-LsdA | | Descriptor: | FE (III) ION, GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, ... | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
