1ANU
 
 | | COHESIN-2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | COHESIN-2 | | Authors: | Shimon, L.J.W, Yaron, S, Shoham, Y, Lamed, R, Morag, E, Bayer, E.A, Frolow, F. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly.
Structure, 5, 1997
|
|
1IDG
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
8CIQ
 
 | | JzTx-34 toxin peptide | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJP
 
 | | JzTx-34 toxin peptide H18A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJS
 
 | | JzTx-34 toxin peptide W31A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJQ
 
 | | JzTx-34 toxin peptide E20A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJR
 
 | | JzTx-34 toxin peptide W25A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJT
 
 | | JzTx-34 toxin peptide W33A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
5ICZ
 
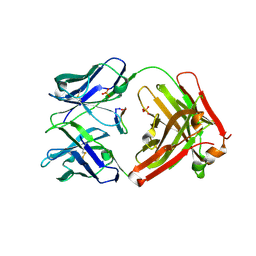 | | Cetuximab Fab in complex with GQFDLSTRRLKG peptide | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, Meditope, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1TQH
 
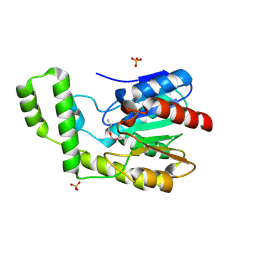 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
7SWT
 
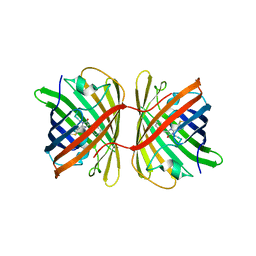 | | Crystal structure of the chromoprotein eforRED | | Descriptor: | Chromoprotein eforRED | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWS
 
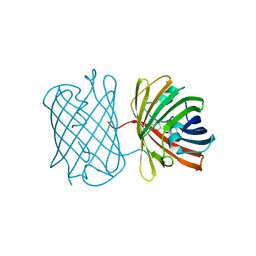 | | Crystal structure of the chromoprotein amilCP | | Descriptor: | BROMIDE ION, CHLORIDE ION, Chromoprotein amilCP | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWR
 
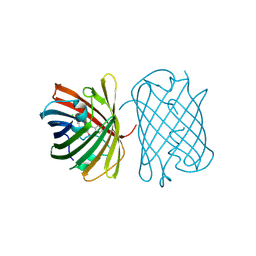 | | Crystal structure of the chromoprotein gfasPurple | | Descriptor: | CHLORIDE ION, Chromoprotein gfasPurple | | Authors: | Caputo, A.T, Newman, J, Peat, T.S, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWU
 
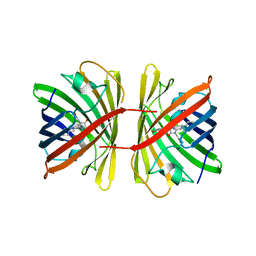 | | Crystal structure of the chromoprotein spisPINK | | Descriptor: | Chromoprotein spisPINK | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6MT1
 
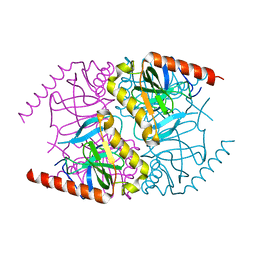 | |
1PPE
 
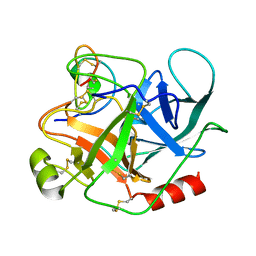 | |
6MT2
 
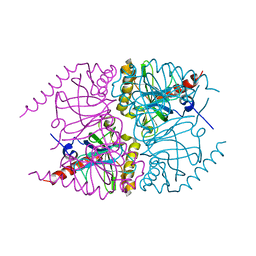 | |
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
2QD5
 
 | | Structure of wild type human ferrochelatase in complex with a lead-porphyrin compound | | Descriptor: | ACETIC ACID, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Meldock, A.E, Dailey, T.A, Ross, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2007-06-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pi-Helix Switch Selective for Porphyrin Deprotonation and Product Release in Human Ferrochelatase.
J.Mol.Biol., 373, 2007
|
|
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
5BTS
 
 | |
7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7JRI
 
 | |
1T66
 
 | | The structure of FAB with intermediate affinity for fluorescein. | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, immunoglobulin heavy chain, immunoglobulin light chain | | Authors: | Terzyan, S, Ramsland, P.A, Voss Jr, E.W, Herron, J.N, Edmundson, A.B. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional Structures of Idiotypically Related Fabs with Intermediate and High Affinity for Fluorescein.
J.Mol.Biol., 339, 2004
|
|
