6YP6
 
 | |
2AYW
 
 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7RWG
 
 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
8PGP
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3193 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanylazetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-cyano-4-(methylsulfonylamino)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3193
To Be Published
|
|
1F94
 
 | | THE 0.97 RESOLUTION STRUCTURE OF BUCANDIN, A NOVEL TOXIN ISOLATED FROM THE MALAYAN KRAIT | | Descriptor: | BUCANDIN | | Authors: | Kuhn, P, Deacon, A.M, Comoso, S, Rajaseger, G, Kini, R.M, Uson, I, Kolatkar, P.R. | | Deposit date: | 2000-07-06 | | Release date: | 2000-07-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The atomic resolution structure of bucandin, a novel toxin isolated from the Malayan krait, determined by direct methods.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5SP2
 
 | |
6F81
 
 | | AKR1B1 at 0.75 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
5XQV
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20L mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6RGP
 
 | | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Second structure of the series with 165 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2GEW
 
 | |
2BF6
 
 | |
4KXV
 
 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
3VII
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6TD8
 
 | | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid) | | Descriptor: | Aldo-keto reductase family 1 member B1, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid)
To Be Published
|
|
4MTU
 
 | | beta-Alanyl-CoA:Ammonia Lyase from Clostridium propionicum | | Descriptor: | Beta-alanyl-CoA:ammonia lyase 2, SULFATE ION, ZINC ION | | Authors: | Heine, A, Reuter, K. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High resolution crystal structure of Clostridium propionicum beta-alanyl-CoA:ammonia lyase, a new member of the "hot dog fold" protein superfamily.
Proteins, 82, 2014
|
|
4Q4G
 
 | |
6FJN
 
 | | Adenovirus species 26 knob protein, 0.97A | | Descriptor: | 1,2-ETHANEDIOL, Fiber | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
6SYE
 
 | |
4PNO
 
 | |
8A3H
 
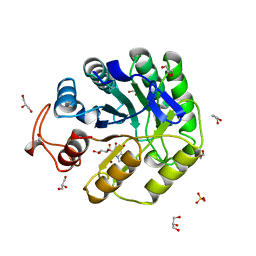 | | Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, ACETATE ION, GLYCEROL, ... | | Authors: | Varrot, A, Schulein, M, Pipelier, M, Vasella, A, Davies, G.J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lateral Protonation of a Glycosidase Inhibitor. Structure of the Bacillus agaradhaerens Cel5A in Complex with a Cellobiose-Derived Imidazole at 0.97 A Resolution
J.Am.Chem.Soc., 121, 1999
|
|
1ZLB
 
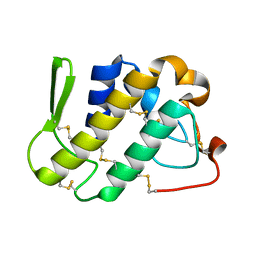 | | Crystal structure of catalytically-active phospholipase A2 in the absence of calcium | | Descriptor: | hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
2WNA
 
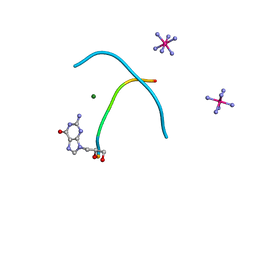 | |
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
485D
 
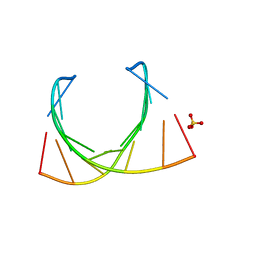 | |
5RC2
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E11b | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
