2PDZ
 
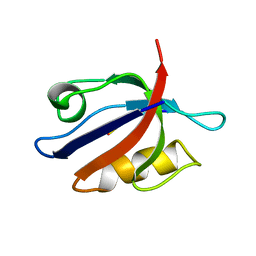 | | SOLUTION STRUCTURE OF THE SYNTROPHIN PDZ DOMAIN IN COMPLEX WITH THE PEPTIDE GVKESLV, NMR, 15 STRUCTURES | | Descriptor: | PEPTIDE GVKESLV, SYNTROPHIN | | Authors: | Schultz, J, Hoffmueller, U, Ashurst, J, Krause, G, Schmieder, P, Macias, M, Schneider-Mergener, J, Oschkinat, H. | | Deposit date: | 1997-12-10 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels.
Nat.Struct.Biol., 5, 1998
|
|
2NDP
 
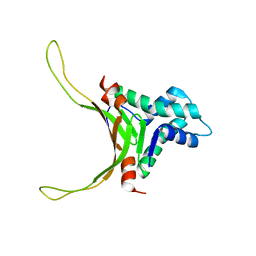 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | Descriptor: | Histone-like DNA-binding superfamily protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | Deposit date: | 2016-09-13 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
2ND4
 
 | |
2ND3
 
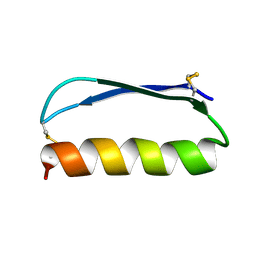 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
201D
 
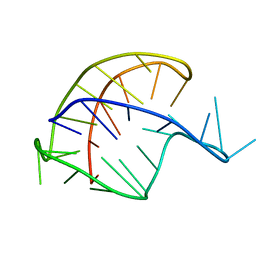 | |
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2MYW
 
 | |
2MPS
 
 | |
2MR7
 
 | |
1WCO
 
 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
2JTG
 
 | | Solution structure of the THAP-zinc finger of THAP1 | | Descriptor: | THAP domain-containing protein 1, ZINC ION | | Authors: | Bessiere, D, Campagne, S, Milon, A, Gervais, V. | | Deposit date: | 2007-07-31 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways
J.Biol.Chem., 283, 2008
|
|
2JT4
 
 | |
2JTS
 
 | | rhodanese with anions from E. coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
2K3N
 
 | |
2K3O
 
 | |
2CCX
 
 | |
2IUE
 
 | |
2I7K
 
 | | Solution Structure of the Bromodomain of Human BRD7 Protein | | Descriptor: | Bromodomain-containing protein 7 | | Authors: | Sun, H, Liu, J, Zhang, J, Huang, H, Wu, J, Shi, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2BTH
 
 | |
2ILX
 
 | |
2ARG
 
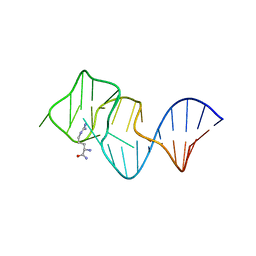 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | Descriptor: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | Authors: | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-08-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
2KM2
 
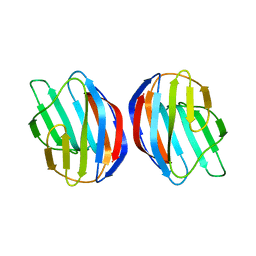 | | Galectin-1 dimer | | Descriptor: | Galectin-1 | | Authors: | Nesmelova, I.V, Ermakova, E, Daragan, V.A, Pang, M, Baum, L.G, Mayo, K.H. | | Deposit date: | 2009-07-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity.
J.Mol.Biol., 397, 2010
|
|
2I3E
 
 | |
2JTQ
 
 | | Rhodanese from E.coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
