1D81
 
 | |
1STP
 
 | |
6GPB
 
 | |
1HSA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HLA-B27 AT 2.1 ANGSTROMS RESOLUTION SUGGESTS A GENERAL MECHANISM FOR TIGHT PEPTIDE BINDING TO MHC | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-B*2705), MODEL PEPTIDE SEQUENCE - ARAAAAAAA | | Authors: | Madden, D.R, Gorga, J.C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1992-08-11 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC.
Cell(Cambridge,Mass.), 70, 1992
|
|
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
1D82
 
 | |
1MEE
 
 | | THE COMPLEX BETWEEN THE SUBTILISIN FROM A MESOPHILIC BACTERIUM AND THE LEECH INHIBITOR EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, MESENTERICOPEPTIDASE | | Authors: | Dauter, Z, Betzel, C, Wilson, K.S. | | Deposit date: | 1991-04-15 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex between the subtilisin from a mesophilic bacterium and the leech inhibitor eglin-C.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5ABP
 
 | |
4GPB
 
 | |
1FAI
 
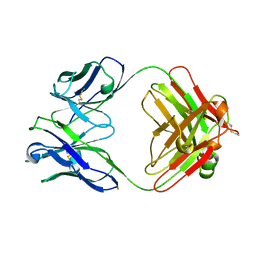 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2GPB
 
 | |
1LPE
 
 | |
2ZTA
 
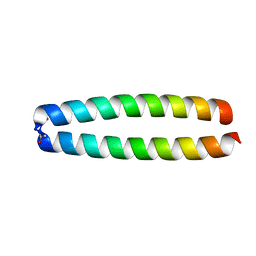 | | X-RAY STRUCTURE OF THE GCN4 LEUCINE ZIPPER, A TWO-STRANDED, PARALLEL COILED COIL | | Descriptor: | GCN4 LEUCINE ZIPPER | | Authors: | O'Shea, E.K, Klemm, J.D, Kim, P.S, Alber, T. | | Deposit date: | 1991-07-05 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil.
Science, 254, 1991
|
|
3IL8
 
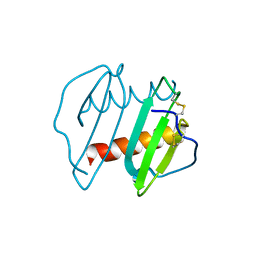 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
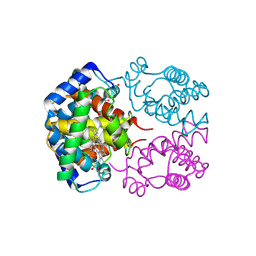
1PBX
 
 | |
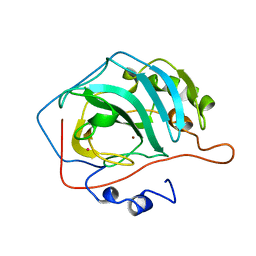
5CA2
 
 | |
4CA2
 
 | |
9RUB
 
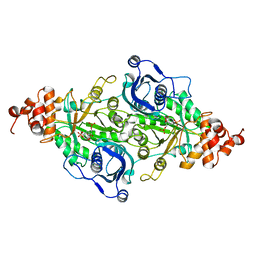 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
9RNT
 
 | |
9LPR
 
 | |
5LPR
 
 | |
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
6RNT
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ADENOSINE 2'-MONOPHOSPHATE AT 1.8-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Ding, J, Koellner, G, Grunert, H.-P, Saenger, W. | | Deposit date: | 1991-08-20 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 complexed with adenosine 2'-monophosphate at 1.8-A resolution.
J.Biol.Chem., 266, 1991
|
|
2LPR
 
 | |
1RGK
 
 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
