1CU0
 
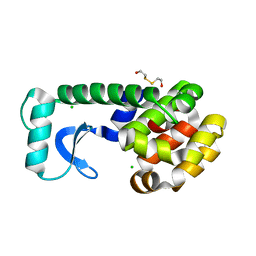 | | T4 LYSOZYME MUTANT I78M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV4
 
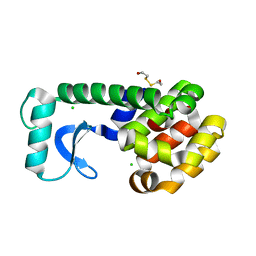 | | T4 LYSOZYME MUTANT L118M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CVK
 
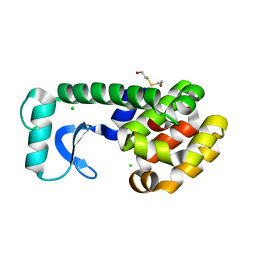 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CU2
 
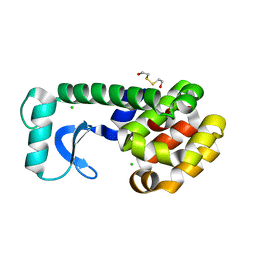 | | T4 LYSOZYME MUTANT L84M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CUQ
 
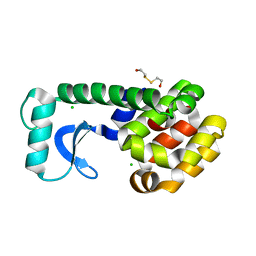 | | T4 LYSOZYME MUTANT V103M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV0
 
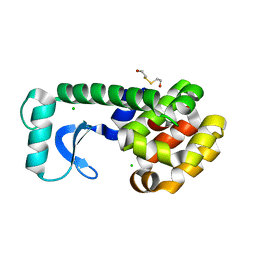 | | T4 LYSOZYME MUTANT F104M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CTW
 
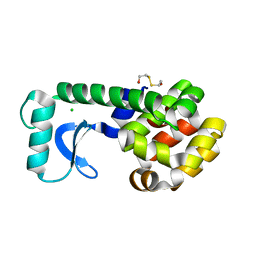 | | T4 LYSOZYME MUTANT I78A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CU5
 
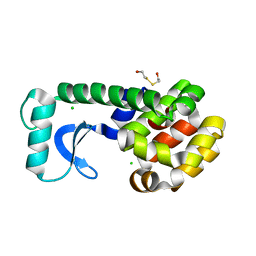 | | T4 LYSOZYME MUTANT L91M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1D8C
 
 | | MALATE SYNTHASE G COMPLEXED WITH MAGNESIUM AND GLYOXYLATE | | Descriptor: | GLYOXYLIC ACID, MAGNESIUM ION, MALATE SYNTHASE G, ... | | Authors: | Howard, B.R, Endrizzi, J.A, Remington, S.J. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 A resolution: mechanistic implications.
Biochemistry, 39, 2000
|
|
1QRR
 
 | | CRYSTAL STRUCTURE OF SQD1 PROTEIN COMPLEX WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Mulichak, A.M, Theisen, M.J, Essigmann, B, Benning, C, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QHA
 
 | | HUMAN HEXOKINASE TYPE I COMPLEXED WITH ATP ANALOGUE AMP-PNP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rosano, C, Sabini, E, Deriu, D, Magnani, M, Bolognesi, M. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of non-catalytic ATP to human hexokinase I highlights the structural components for enzyme-membrane association control.
Structure Fold.Des., 7, 1999
|
|
1QHL
 
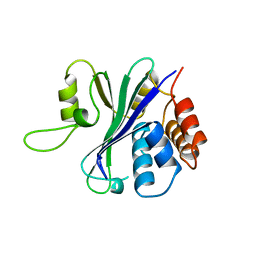 | |
1QSZ
 
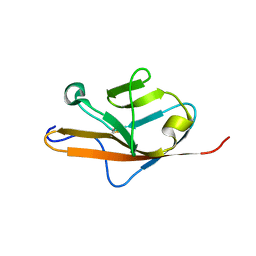 | | THE VEGF-BINDING DOMAIN OF FLT-1 (MINIMIZED MEAN) | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Starovasnik, M.A, Christinger, H.W, Wiesmann, C, Champe, M.A, de Vos, A.M, Skelton, N.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-11-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VEGF-binding domain of Flt-1: comparison of its free and bound states.
J.Mol.Biol., 293, 1999
|
|
1QCZ
 
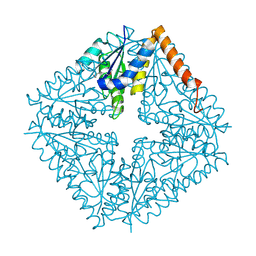 | |
2NLR
 
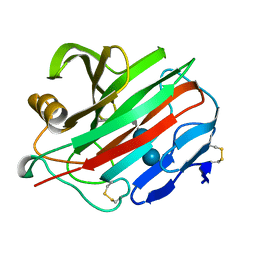 | |
1QC0
 
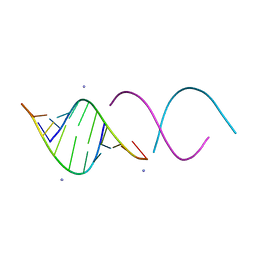 | | CRYSTAL STRUCTURE OF A 19 BASE PAIR COPY CONTROL RELATED RNA DUPLEX | | Descriptor: | 5'-R(*GP*CP*AP*CP*CP*GP*CP*UP*AP*CP*CP*AP*AP*CP*GP*GP*UP*GP*C)-3', 5'-R(*GP*CP*AP*CP*CP*GP*UP*UP*GP*GP*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', 5'-R(*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', ... | | Authors: | Klosterman, P.S, Shah, S.A, Steitz, T.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two plasmid copy control related RNA duplexes: An 18 base pair duplex at 1.20 A resolution and a 19 base pair duplex at 1.55 A resolution.
Biochemistry, 38, 1999
|
|
1QCU
 
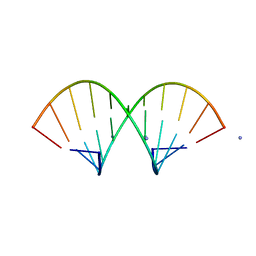 | |
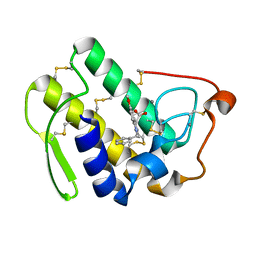
1DCY
 
 | |
1DB4
 
 | | HUMAN S-PLA2 IN COMPLEX WITH INDOLE 8 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, [3-(1-BENZYL-3-CARBAMOYLMETHYL-2-METHYL-1H-INDOL-5-YLOXY)-PROPYL-]-PHOSPHONIC ACID | | Authors: | Chirgadze, N.Y, Schevitz, R.W, Wery, J.-P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of the first potent and selective inhibitor of human non-pancreatic secretory phospholipase A2.
Nat.Struct.Biol., 2, 1995
|
|
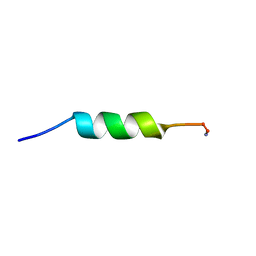
1D9L
 
 | |
1CXW
 
 | | THE SECOND TYPE II MODULE FROM HUMAN MATRIX METALLOPROTEINASE 2 | | Descriptor: | HUMAN MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Grishaev, A, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-12 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | The second type II module from human matrix metalloproteinase 2: structure, function and dynamics.
Structure Fold.Des., 7, 1999
|
|
1D9J
 
 | |
1D9P
 
 | |
1DB5
 
 | | HUMAN S-PLA2 IN COMPLEX WITH INDOLE 6 | | Descriptor: | 4-(1-BENZYL-3-CARBAMOYLMETHYL-2-METHYL-1H-INDOL-5-YLOXY)-BUTYRIC ACID, CALCIUM ION, PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Chirgadze, N.Y, Schevitz, R.W, Wery, J.-P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-12 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of the first potent and selective inhibitor of human non-pancreatic secretory phospholipase A2.
Nat.Struct.Biol., 2, 1995
|
|
