8F7T
 
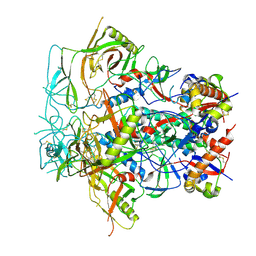 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
2OTO
 
 | |
3O6K
 
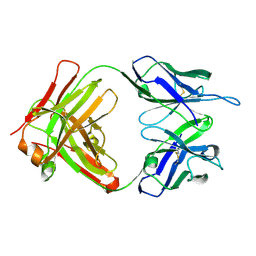 | | Crystal structure of anti-Tat HIV Fab'11H6H1 | | Descriptor: | 11H6H1 Fab' heavy chain, 11H6H1 Fab' light chain | | Authors: | Serriere, J, Gouet, P, Guillon, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fab'-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography.
J.Mol.Biol., 405, 2011
|
|
2NMS
 
 | |
2P7R
 
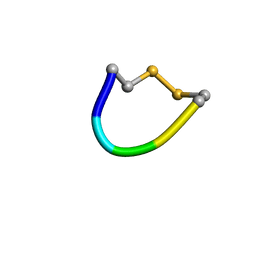 | | Cyclic pentapeptide which inhibits Hantavirus | | Descriptor: | cyclo-CPFVC | | Authors: | Hall, P.R, Malone, L, Sillerud, L.O, Ye, C, Hjelle, B, Larson, R.S. | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Characterization and NMR solution structure of a novel cyclic pentapeptide inhibitor of pathogenic hantaviruses
CHEM.BIOL.DRUG DES., 69, 2007
|
|
2P28
 
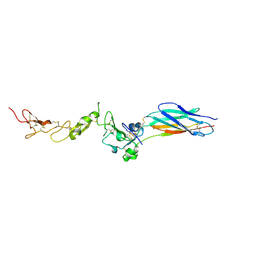 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
3M6F
 
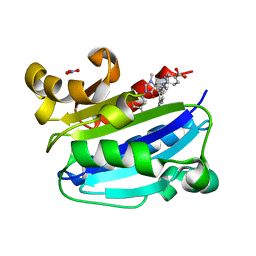 | | CD11A I-domain complexed with 6-((5S,9R)-9-(4-CYANOPHENYL)-3-(3,5-DICHLOROPHENYL)-1-METHYL-2,4-DIOXO-1,3,7- TRIAZASPIRO[4.4]NON-7-YL)NICOTINIC ACID | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, NITRATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small molecule antagonist of leukocyte function associated antigen-1 (LFA-1): structure-activity relationships leading to the identification of 6-((5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]nonan-7-yl)nicotinic acid (BMS-688521).
J.Med.Chem., 53, 2010
|
|
3O6L
 
 | |
2P26
 
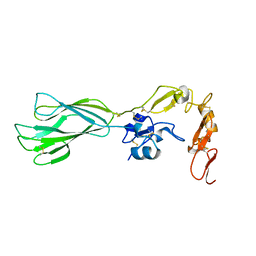 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-06 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
2NOJ
 
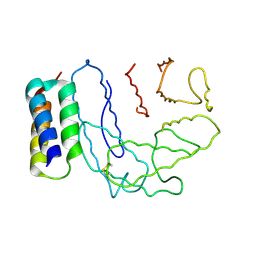 | | Crystal structure of Ehp / C3d complex | | Descriptor: | Complement C3, Efb homologous protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Ehp, a Secreted Complement Inhibitory Protein from Staphylococcus aureus.
J.Biol.Chem., 282, 2007
|
|
2Q2O
 
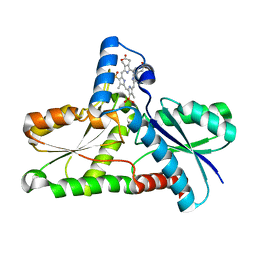 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q3J
 
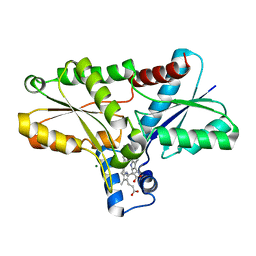 | | Crystal structure of the His183Ala variant of Bacillus subtilis ferrochelatase in complex with N-Methyl Mesoporphyrin | | Descriptor: | Ferrochelatase, MAGNESIUM ION, N-METHYL PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-30 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2MUD
 
 | |
2MG1
 
 | | NMR assignment and structure of a peptide derived from the trans-membrane region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Apellaniz, B, Huarte, N.L, Nieva, J.L, Jimenez, M.A. | | Deposit date: | 2013-10-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
2PY0
 
 | | Crystal structure of Cs1 pilin chimera | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fimbrial protein | | Authors: | Kao, D.J, Churchill, M.E, Hodges, R.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Animal Protection and Structural Studies of a Consensus Sequence Vaccine Targeting the Receptor Binding Domain of the Type IV Pilus of Pseudomonas aeruginosa.
J.Mol.Biol., 374, 2007
|
|
2Q2N
 
 | | Crystal structure of Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2QA0
 
 | | Structure of Adeno-Associated virus serotype 8 | | Descriptor: | Capsid protein, SODIUM ION | | Authors: | Nam, H.-J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of adeno-associated virus serotype 8, a gene therapy vector.
J.Virol., 81, 2007
|
|
6RPJ
 
 | |
6RU5
 
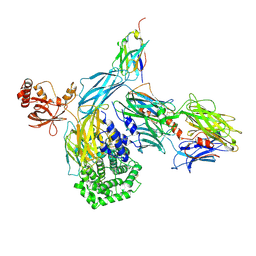 | | human complement C3 in complex with the hC3Nb1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
2BLP
 
 | | RNase before unattenuated X-RAY burn | | Descriptor: | CHLORIDE ION, RIBONUCLEASE PANCREATIC PRECURSOR | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving radiation-damage substructures for RIP.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
8UG9
 
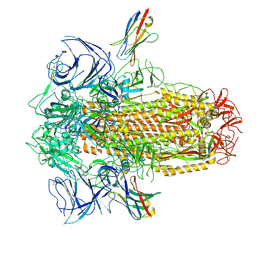 | | XBB.1.5 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
2BHI
 
 | | Crystal structure of Taiwan cobra cardiotoxin A3 complexed with sulfogalactoceramide | | Descriptor: | CYTOTOXIN 3, HEXAETHYLENE GLYCOL MONODECYL ETHER, SULFOGALACTOCERAMIDE | | Authors: | Wang, C.-H, Liu, J.-H, Wu, P.-L, Lee, S.-C, Hsiao, C.-D, Wu, W.-G. | | Deposit date: | 2005-01-12 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycosphingolipid-Facilitated Membrane Insertion and Internalization of Cobra Cardiotoxin: The Sulfatide/Cardiotoxin Complex Structure in a Membrane-Like Environment Suggests a Lipid-Dependent Cell-Penetrating Mechanism for Membrane Binding Polypeptides.
J.Biol.Chem., 281, 2006
|
|
2C4F
 
 | | crystal structure of factor VII.stf complexed with pd0297121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[6-{3-[AMINO(IMINO)METHYL]PHENOXY}-4-(DIISOPROPYLAMINO)-3,5-DIFLUOROPYRIDIN-2-YL]OXY}-5-[(ISOBUTYLAMINO)CARBONYL]BEN ZOIC ACID, CALCIUM ION, ... | | Authors: | Kohrt, J.T, Zhang, E. | | Deposit date: | 2005-10-18 | | Release date: | 2006-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of Fluoropyridine-Based Inhibitors of the Factor Viia/Tf Complex--Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ALY
 
 | |
2CDG
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
