6NMW
 
 | |
4GPT
 
 | | Crystal structure of KPT251 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}ethyl)-1,3,4-oxadiazole, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells.
Leukemia, 27, 2013
|
|
3NCT
 
 | | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of reca | | Descriptor: | Protein psiB | | Authors: | Petrova, V, Satyshur, K.A, George, N.P, McCaslin, D, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of RecA.
J.Biol.Chem., 285, 2010
|
|
6N89
 
 | |
6MZL
 
 | | Human TFIID canonical state | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
4HVU
 
 | |
6MDP
 
 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
4J0U
 
 | | Crystal structure of IFIT5/ISG58 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Liu, Y, Liang, H, Feng, F, Yuan, L, Wang, Y.E, Crowley, C, Lv, Z, Li, J, Zeng, S, Cheng, G. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal Structure of IFIT5
To be Published
|
|
3OCH
 
 | | Chemically Self-assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic | | Descriptor: | (2S,2'S)-5,5'-(nonane-1,9-diyldiimino)bis(2-{[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)amino}phenyl)carbonyl]amino}-5-oxopentanoic acid) (non-preferred name), CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cody, V. | | Deposit date: | 2010-08-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Chemically Self-Assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic.
J.Am.Chem.Soc., 132, 2010
|
|
3MYH
 
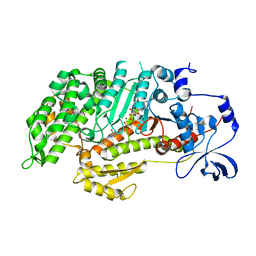 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
3MYL
 
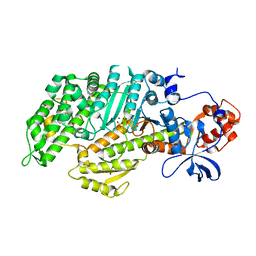 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | MAGNESIUM ION, Myosin-2 heavy chain, PYROPHOSPHATE 2- | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
5OTE
 
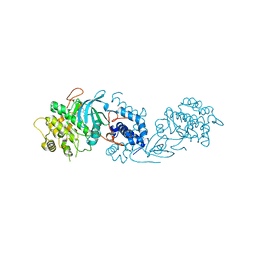 | | MRCK beta in complex with BDP-00008900 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(6~{S})-1,8-diazaspiro[5.5]undecan-8-yl]-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-1,3-thiazole, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Potent and Selective MRCK Inhibitors with Therapeutic Effect on Skin Cancer.
Cancer Res., 78, 2018
|
|
4HAX
 
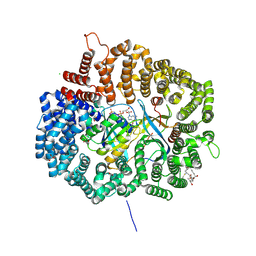 | |
4HB0
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAV
 
 | |
3MYK
 
 | | Insights into the Importance of Hydrogen Bonding in the Gamma-Phosphate Binding Pocket of Myosin: Structural and Functional Studies of Ser236 | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, MAGNESIUM ION, Myosin-2 heavy chain, ... | | Authors: | Frye, J.J, Klenchin, V.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 2010-05-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into the importance of hydrogen bonding in the gamma-phosphate binding pocket of myosin: structural and functional studies of serine 236
Biochemistry, 49, 2010
|
|
4FWK
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
4FWQ
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
4FWO
 
 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HAT
 
 | |
3LQQ
 
 | | Structure of the CED-4 Apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.534 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
5VFP
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VHN
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VFR
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VHJ
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
