3VSZ
 
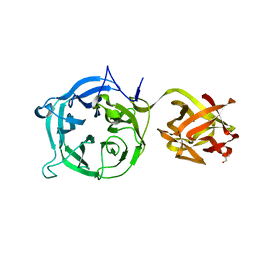 | | Crystal structure of Ct1,3Gal43A in complex with galactan | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
3CSF
 
 | |
4QJ0
 
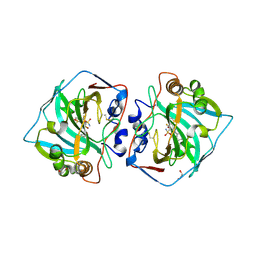 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
3VWP
 
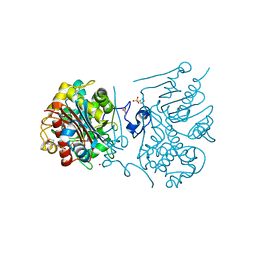 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VTX
 
 | | Crystal structure of MamA protein | | Descriptor: | GLYCEROL, MamA | | Authors: | Zeytuni, N, Baran, D, Davidov, G, Zarivach, R. | | Deposit date: | 2012-06-08 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA
J.Struct.Biol., 180, 2012
|
|
3CXD
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Yang, H, Zhong, C, Ding, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3CXU
 
 | | Structure of a Y149F mutant of epoxide hydrolase from Solanum tuberosum | | Descriptor: | Epoxide hydrolase, TETRAETHYLENE GLYCOL | | Authors: | Naworyta, A, Mowbray, S.L, Widersten, M, Thomaeus, A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability
Protein Sci., 17, 2008
|
|
4QNQ
 
 | |
3W1H
 
 | |
4QOL
 
 | | Structure of Bacillus pumilus catalase | | Descriptor: | ACETATE ION, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
3F00
 
 | |
3VSW
 
 | | Human renin in complex with compound 8 | | Descriptor: | (2S,4S,5S)-5-amino-N-(3-amino-2,2-dimethyl-3-oxopropyl)-4-hydroxy-6-{4-[2-(3-methoxypropoxy)phenyl]-3-oxopiperazin-1-yl}-2-(propan-2-yl)hexanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-05-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and optimization of novel (2S,4S,5S)-5-amino-6-(2,2-dimethyl-5-oxo-4-phenylpiperazin-1-yl)-4-hydroxy-2-isopropylhexanamides as renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3FNQ
 
 | | Crystal structure of schistosoma purine nucleoside phosphorylase in complex with hypoxanthine | | Descriptor: | DIMETHYL SULFOXIDE, HYPOXANTHINE, Purine-nucleoside phosphorylase, ... | | Authors: | Castilho, M.S, Pereira, H.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-12-26 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3F0F
 
 | |
4QJP
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-phenylethyl)sulfonyl]benzenesulfonamide, CITRIC ACID, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-06-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
4QL1
 
 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
3W13
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alphact peptide(693-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowski, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
3VUC
 
 | | Human renin in complex with compound 5 | | Descriptor: | (2R,4S,5S)-5-amino-6-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-2-ethyl-4-hydroxy-N-[(1R,2S,3S,5S,7S)-5-hydroxytricyclo[3.3.1.1~3,7~]dec-2-yl]hexanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of DS-8108b, a Novel Orally Bioavailable Renin Inhibitor.
Acs Med.Chem.Lett., 3, 2012
|
|
3VUX
 
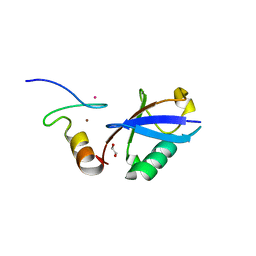 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form II | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Polyubiquitin-C, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3VWN
 
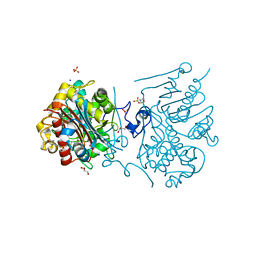 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VVN
 
 | |
4QN1
 
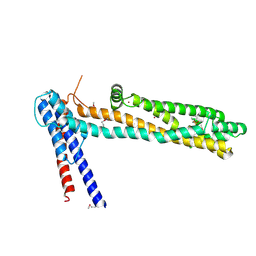 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
4QOQ
 
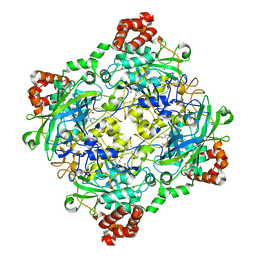 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QIZ
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 13, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
