1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1OCZ
 
 | |
1FCE
 
 | | PROCESSIVE ENDOCELLULASE CELF OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, CELLULASE CELF, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Parsiegla, G, Juy, M, Haser, R. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 A resolution.
EMBO J., 17, 1998
|
|
1BI9
 
 | | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, RETINAL DEHYDROGENASE TYPE II | | Authors: | Newcomer, M.E, Lamb, A.L. | | Deposit date: | 1998-06-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity.
Biochemistry, 38, 1999
|
|
1BWM
 
 | | A SINGLE-CHAIN T CELL RECEPTOR | | Descriptor: | PROTEIN (ALPHA-BETA T CELL RECEPTOR (TCR) (D10)) | | Authors: | Hare, B.J, Wyss, D.F, Reinherz, E.L, Wagner, G. | | Deposit date: | 1998-09-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor.
Nat.Struct.Biol., 6, 1999
|
|
4DCG
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT D182A COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CNO
 
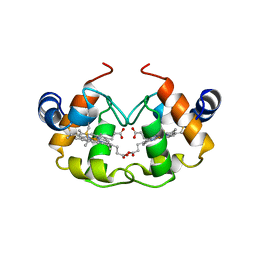 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
1BKG
 
 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS WITH MALEATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-07-07 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1BKY
 
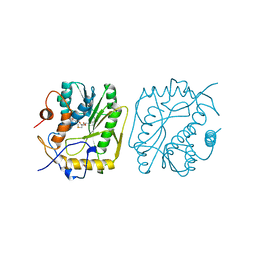 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 1-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BJF
 
 | |
1BJW
 
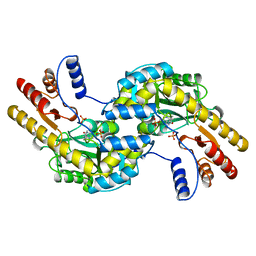 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
3ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1QTD
 
 | |
1XPA
 
 | | SOLUTION STRUCTURE OF THE DNA-AND RPA-BINDING DOMAIN OF THE HUMAN REPAIR FACTOR XPA, NMR, 1 STRUCTURE | | Descriptor: | XPA, ZINC ION | | Authors: | Ikegami, T, Kuraoka, I, Saijo, M, Kodo, N, Kyogoku, Y, Morikawa, K, Tanaka, K, Shirakawa, M. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA.
Nat.Struct.Biol., 5, 1998
|
|
1QTH
 
 | |
1QTC
 
 | |
4ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | Descriptor: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1EQA
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT E233Q COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1MKP
 
 | | CRYSTAL STRUCTURE OF PYST1 (MKP3) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PYST1 | | Authors: | Stewart, A.E, Dowd, S, Keyse, S, Mcdonald, N.Q. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation.
Nat.Struct.Biol., 6, 1999
|
|
2EUH
 
 | | HOLO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE COMPLEX WITH NADP+ | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
1TLG
 
 | | STRUCTURE OF A TUNICATE C-TYPE LECTIN COMPLEXED WITH D-GALACTOSE | | Descriptor: | CALCIUM ION, POLYANDROCARPA LECTIN, ZINC ION, ... | | Authors: | Poget, S.F, Legge, G.B, Bycroft, M, Williams, R.L. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a tunicate C-type lectin from Polyandrocarpa misakiensis complexed with D -galactose.
J.Mol.Biol., 290, 1999
|
|
1BK7
 
 | | RIBONUCLEASE MC1 FROM THE SEEDS OF BITTER GOURD | | Descriptor: | PROTEIN (RIBONUCLEASE MC1) | | Authors: | Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-07-15 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a ribonuclease from the seeds of bitter gourd (Momordica charantia) at 1.75 A resolution.
Biochim.Biophys.Acta, 1433, 1999
|
|
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1B65
 
 | | Structure of l-aminopeptidase d-ala-esterase/amidase from ochrobactrum anthropi, a prototype for the serine aminopeptidases, reveals a new variant among the ntn hydrolase fold | | Descriptor: | PROTEIN (AMINOPEPTIDASE) | | Authors: | Bompard-Gilles, C, Villeret, V, Davies, G.J, Fanuel, L, Joris, B, Frere, J.M, Van Beeumen, J. | | Deposit date: | 1999-01-20 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new variant of the Ntn hydrolase fold revealed by the crystal structure
of L-aminopeptidase D-ala-esterase/amidase from Ochrobactrum anthropi.
Structure Fold.Des., 8, 2000
|
|
1BYF
 
 | | STRUCTURE OF TC14; A C-TYPE LECTIN FROM THE TUNICATE POLYANDROCARPA MISAKIENSIS | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Poget, S.F, Legge, G.B, Bycroft, M, Williams, R.L. | | Deposit date: | 1998-10-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a tunicate C-type lectin from Polyandrocarpa misakiensis complexed with D -galactose.
J.Mol.Biol., 290, 1999
|
|
