5RZ0
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1497321453 | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)(pyrrolidin-1-yl)methanone, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
8HAO
 
 | | Human parathyroid hormone receptor-1 dimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
5RZH
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434890 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-N-phenylpiperazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZV
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z31504642 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-phenylacetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
8HDO
 
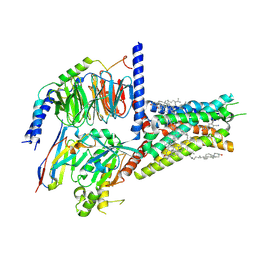 | | Structure of A2BR bound to synthetic agonists BAY 60-6583 | | Descriptor: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
5KZT
 
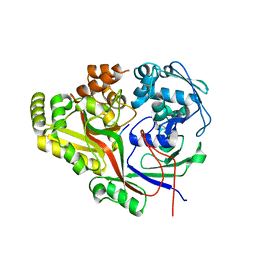 | | Listeria monocytogenes OppA bound to peptide | | Descriptor: | Hexamer peptide: SER-ASP-GLU-SER-LYS-GLY, Hexamer peptide: SER-ASP-GLU-SER-SER-GLY, Peptide/nickel transport system substrate-binding protein | | Authors: | Light, S.H, Whiteley, A.T, Minasov, G, Portnoy, D.A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Listeria monocytogenes OppA bound to peptide
To Be Published
|
|
8CE8
 
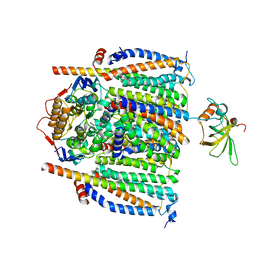 | | Cytochrome c maturation complex CcmABCDE | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Cytochrome c-type biogenesis protein CcmE, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7VBH
 
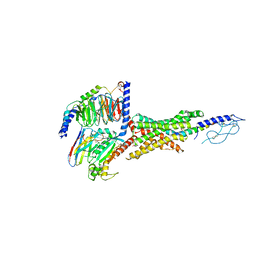 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
4XMV
 
 | |
7D63
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
4XN1
 
 | |
4XNB
 
 | |
4XO5
 
 | |
5L3P
 
 | |
4XP9
 
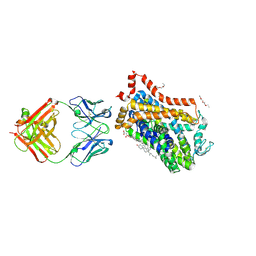 | | X-ray structure of Drosophila dopamine transporter bound to psychostimulant D-amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1MF8
 
 | |
4XRG
 
 | |
8HDP
 
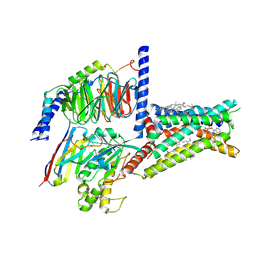 | | Structure of A2BR bound to endogenous agonists adenosine | | Descriptor: | ADENOSINE, Adenosine A2b receptor, CHOLESTEROL, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of adenosine receptor A 2B R bound to endogenous and synthetic agonists.
Cell Discov, 8, 2022
|
|
8H8J
 
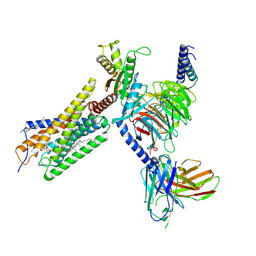 | | Lodoxamide-bound GPR35 in complex with G13 | | Descriptor: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | Authors: | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-10-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
4XZ6
 
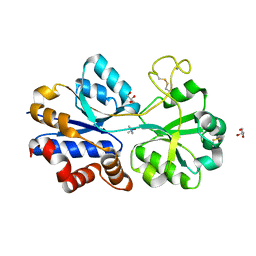 | | TmoX in complex with TMAO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
4XZB
 
 | |
4XZW
 
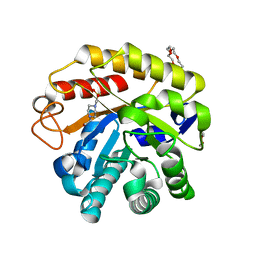 | | Endo-glucanase chimera C10 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Lee, C.C, Chang, C.J, Ho, T.H.D, Chao, Y.C, Wang, A.H.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Endo-glucanase chimera C10
To Be Published
|
|
5RZ8
 
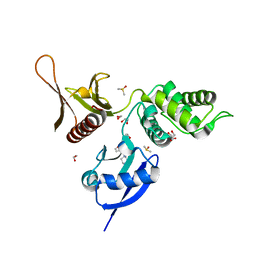 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1263714198 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1,2-oxazol-3-yl)methyl]piperazine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZO
 
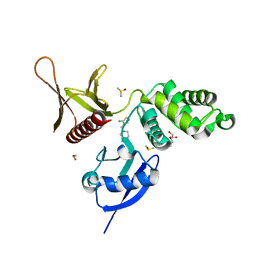 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434925 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5-chloro-1,2,3-thiadiazol-4-yl)methyl]morpholine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
