8H8R
 
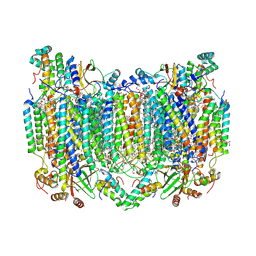 | | Bovine Heart Cytochrome c Oxidase in the Calcium-bound Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Calcium-bound structure of bovine cytochrome c oxidase.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
1OCO
 
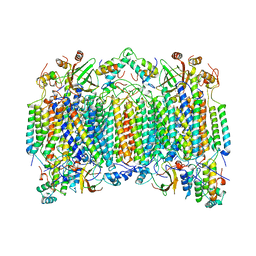 | |
1OCR
 
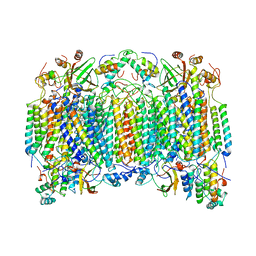 | |
1OCZ
 
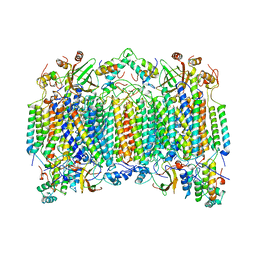 | |
1QLE
 
 | |
3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OMI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1OCC
 
 | | STRUCTURE OF BOVINE HEART CYTOCHROME C OXIDASE AT THE FULLY OXIDIZED STATE | | Descriptor: | COPPER (II) ION, CYTOCHROME C OXIDASE, HEME-A, ... | | Authors: | Tsukihara, T, Aoyama, H, Yamashita, E, Tomizaki, T, Yamaguchi, H, Shinzawa-Itoh, K, Nakashima, R, Yaono, R, Yoshikawa, S. | | Deposit date: | 1996-04-18 | | Release date: | 1996-12-07 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A.
Science, 272, 1996
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8IJN
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6KOE
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOC
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8D4T
 
 | | Mammalian CIV with GDN bound | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Di Trani, J, Rubinstein, J. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian complex IV inhibition by steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6KOB
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | Deposit date: | 2019-10-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
3WG7
 
 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
6L3H
 
 | | Cryo-EM structure of dimeric quinol dependent Nitric Oxide Reductase (qNOR) from the pathogen Neisseria meninigitidis | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Gopalasingam, C.C, Johnson, R.M, Tosha, T, Muench, S.P, Muramoto, K, Antonyuk, S.V, Shiro, Y, Hasnain, S.S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8K6Y
 
 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8DH6
 
 | | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1, ... | | Authors: | Godoy, A.S, Song, Y, Cheruvara, H, Quigley, A, Oliva, G. | | Deposit date: | 2022-06-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs
To Be Published
|
|
6YMY
 
 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
8F6C
 
 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
