1PL3
 
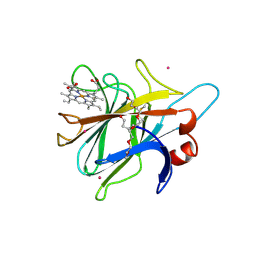 | | Cytochrome Domain Of Cellobiose Dehydrogenase, M65H mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Rotsaert, F.A.J, Hallberg, B.M, de Vries, S, Moenne-Loccoz, P, Divne, C, Gold, M.H. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biophysical and Structural Analysis of a Novel Heme b Iron Ligation in the Flavocytochrome Cellobiose Dehydrogenase.
J.Biol.Chem., 278, 2003
|
|
2WHQ
 
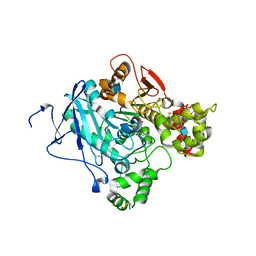 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin (aged) in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
1PN7
 
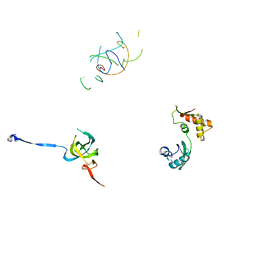 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PKW
 
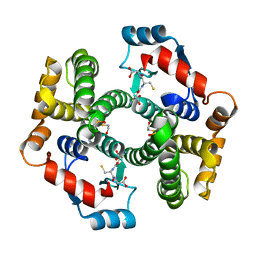 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2WLL
 
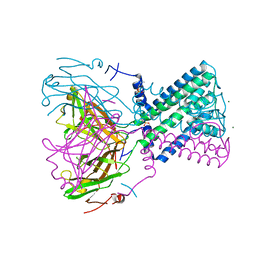 | | POTASSIUM CHANNEL FROM BURKHOLDERIA PSEUDOMALLEI | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, MAGNESIUM ION, POTASSIUM CHANNEL, ... | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, VandenBerg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
1PU6
 
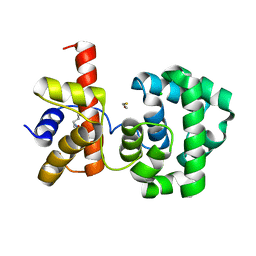 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
2WZO
 
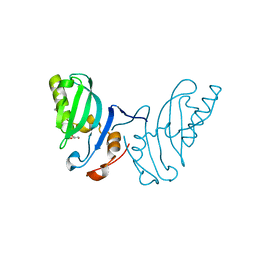 | | The structure of the FYR domain | | Descriptor: | GLYCEROL, TRANSFORMING GROWTH FACTOR BETA REGULATOR 1 | | Authors: | Garcia-Alai, M.M, Allen, M.D, Joerger, A.C, Bycroft, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Fyr Domain of Transforming Growth Factor Beta Regulator 1 (Tbrg1)(Pna)
Protein Sci., 19, 2010
|
|
1PUF
 
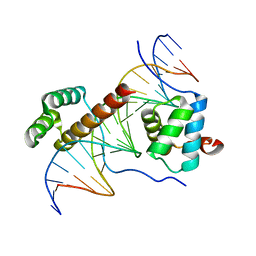 | | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | | Descriptor: | 5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*CP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3', Homeobox protein Hox-A9, ... | | Authors: | Laronde-Leblanc, N.A, Wolberger, C. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE OF HOXA9 AND PBX1 BOUND TO DNA: HOX HEXAPEPTIDE AND DNA RECOGNITION ANTERIOR TO POSTERIOR
Genes Dev., 17, 2003
|
|
3K7A
 
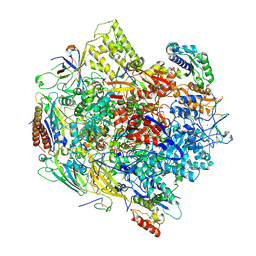 | | Crystal Structure of an RNA polymerase II-TFIIB complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Liu, X, Bushnell, D.A, Wang, D, Calero, G, Kornberg, R.D. | | Deposit date: | 2009-10-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism.
Science, 327, 2010
|
|
2WSB
 
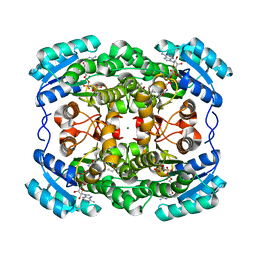 | | Crystal structure of the short-chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD | | Descriptor: | GALACTITOL DEHYDROGENASE, MAGNESIUM ION, N-PROPANOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-09-04 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
1PL1
 
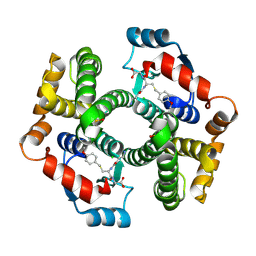 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
1Q2C
 
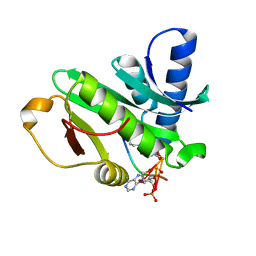 | |
2X04
 
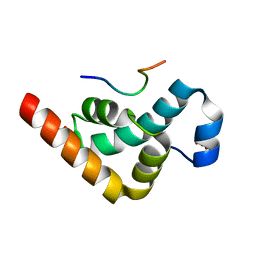 | | Crystal structure of the PABC-TNRC6C complex | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1, SULFATE ION, TRINUCLEOTIDE REPEAT-CONTAINING GENE 6C PROTEIN | | Authors: | Jinek, M, Fabian, M.R, Coyle, S.M, Sonenberg, N, Doudna, J.A. | | Deposit date: | 2009-12-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Insights Into the Human Gw182-Pabc Interaction in Microrna-Mediated Deadenylation
Nat.Struct.Mol.Biol., 17, 2010
|
|
1Z5A
 
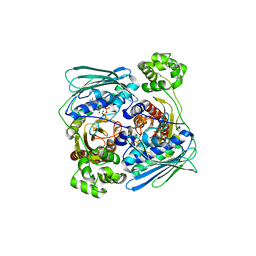 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
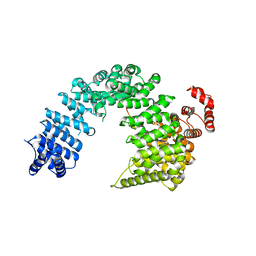
5YVG
 
 | |
3RTS
 
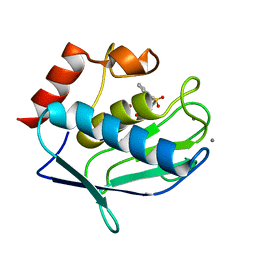 | | Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-[(2-phenylethyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
1PVG
 
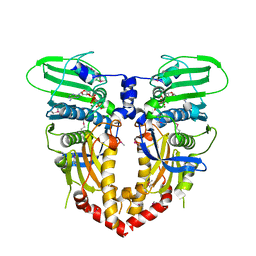 | |
1PU7
 
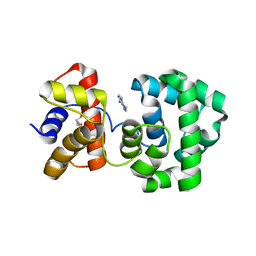 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
3K60
 
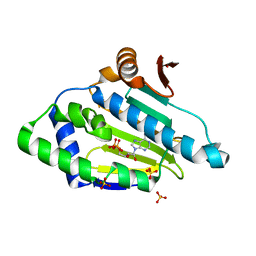 | |
8B7S
 
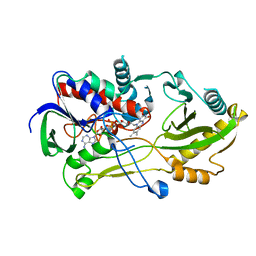 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
6ZJH
 
 | | Trehalose transferase from Thermoproteus uzoniensis soaked with trehalose | | Descriptor: | GLYCEROL, Trehalose phosphorylase/synthase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
3K8Z
 
 | | Crystal Structure of Gudb1 a decryptified secondary glutamate dehydrogenase from B. subtilis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
5WB8
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Epigen, ... | | Authors: | Bessman, N.J, Freed, D.M, Moore, J.O, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
